TFBSPred: A functional transcription factor binding site prediction webtool for humans and mice
- Authors:
- Published online on: September 3, 2021 https://doi.org/10.3892/ije.2021.9
- Article Number: 9
-
Copyright: © Zogopoulos et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
RNA polymerase II binds to the promoter of a gene and it assembles the transcription mechanism by gathering general transcription factors (GTFs), creating the pre-initiation complex to initiate transcription. Transcription is regulated by cis-regulatory elements, including the promoter. Distal elements can exert a positive effect on transcription (termed enhancers) or a negative effect (termed silencers) (1-3). Transcription factors (TFs) are proteins that bind to such regulatory regions of genes recognising multiple specific DNA sequence motifs, termed TF binding sites (TFBSs) (4). There are >1,600 human TFs catalogued (5), controlling processes of cell type specification, developmental patterning (6), as well as specific biological pathways (7). The binding affinity of a TF is dependent on its DNA-binding domain and the specific sequence of nucleotides which is targeted (8). Potential binding sites are predicted based on matches to a consensus sequence, often allowing for certain mismatches. The methods originally used for searching genome sequences to predict TFBSs were based on position weight matrices (PWMs), also known as position-specific scoring matrices (PSSMs) (9), derived from the multiple sequence alignment of experimentally verified DNA target motifs. A weight matrix is a two dimensional array of values that represent the score for finding each of the four nucleotides at each position in the DNA sequence (10). The main databases for the collection of PWMs are TRANSFAC (11) and JASPAR (12). The majority of algorithms used for searching for PWMs in genomic sequences are MATCH (13) and FIMO (14). To simulate TFBS motif intricacies, more complex models have been proposed in recent years (15-18) and, in particular, hidden Markov models (HMMs) (19) have been applied successfully for TFBS prediction (20,21). To account further for TFBS length variability and interactions between nucleotides, HMM-based TF flexible models (TFFMs) (22) were developed, by consulting already existing JASPAR models. They can be graphically represented with a sequence logo, in a manner similar to PWMs (23).
Comparing the relative order of gene orthologs in the human and mouse genomes has revealed that a long-range sequence organisation has been preserved to a large extent from their last common ancestor (24). Approximately 80% of the common genes can be matched between the two organisms with the addition of a high rate of conservation of nucleotides (25). Genes that share close evolutionary associations are likely to possess similar functions and, likewise, functionally similar cis-regulatory elements have been shown to be conserved between species (1,26). Therefore, it can be considered that the majority of functional TFBS sequences are conserved between human and mouse.
DNase I is an endonuclease that cleaves DNA adjacent to pyrimidine nucleotides. In order for DNase I to cleave a DNA strand, it needs to be able to access it. Within cells, TFs displace histone octamers, unwinding the tightly packed chromatin structure. Through the technique of DNase I hypersensitivity (DH) assays (27) and its evolutions (28), DH sites, which denote the open and accessible areas that DNase I can operate on, are discovered. Analysing the whole genome accessibility landscape using DNase I, yields DH maps that denote parts of the genome that are probably transcriptionally active. From these data, it is possible to discover potential cell-type specific TFBSs, as the DH areas originate from open chromatin regions that contain regulatory elements of active genes (29-31).
Predicting TFBSs in regulatory sequences of a gene of interest requires some of the aforementioned computational tools. The present study introduces TFBSPred, a TFBS prediction webtool which eliminates false discoveries by using novel TFFM searches on cell type-specific open chromatin regions, which are conserved between Homo sapiens and Mus musculus.
Data and methods
Data collection
Using in-house PHP scripts, the present study downloaded and processed various Homo sapiens and Mus musculus genomic data, such as gene symbols and annotations from HGNC (32) and MGI (33), as well as gene stable IDs, transcript stable IDs and transcription start site (TSS) information from Ensembl (34) through BioMart (35). To determine the open chromatin regions of each biological replicate of each cell line and/or treatment, BroadPeak (36) DH data (31) for human hg19 and mouse mm9 genomes were downloaded from the UCSC Genome Browser database (37). The LiftOver tool (38) was used to update the DH data coordinates to those of the latest versions of the two genomes (hg38 for human and mm10 for mouse), removing unmapped or duplicate regions. DH data for each replicate, cover, on average, 3.1 and 3.6% of the human and mouse genomes, respectively (Table SI).
Furthermore, ENCODE (39) common cell type details from UCSC Genome Browser were collected and parsed. To determine the conserved sequences between Homo sapiens and Mus musculus, Ensembl Compara (40) pairwise alignments in multiple alignment format (MAF) for the latest versions of the two genomes were downloaded, which were constructed using the LASTZ (41) alignment program. When parsing all MAF files, all pairwise alignments were converted into FASTA format and their genomic coordinates were extracted. The alignments cover, on average, 32.8 and 35.6% of the human and mouse genomes, respectively (Table SII). All aforementioned data were stored in a relational database using the MySQL management system (https://www.mysql.com/) on a Linux Ubuntu 64-bit, 16-core (Intel(R) Xeon(R) CPU E5-2650 v3), 64 GB memory virtual machine, which is provided by GRNET (https://okeanos-knossos.grnet.gr/).
The TF and TFFM data were procured from the latest version of the JASPAR database (http://jaspar.genereg.net/). From the TFFMs available, only the detailed trained ones that belonged to vertebrates were retained. A total of 462 TFFMs are included.
Web interface
The website is hosted on an Apache2 HTTP server. It was developed in HTML5, which was validated by HTML validator addon (https://www.gueury.com/mozilla/). Bootstrap styling library (https://getbootstrap.com/) was used for the website design. JavaScript was used to implement the selection checkboxes and autocompletion fields. All scripts were written in PHP scripting language.
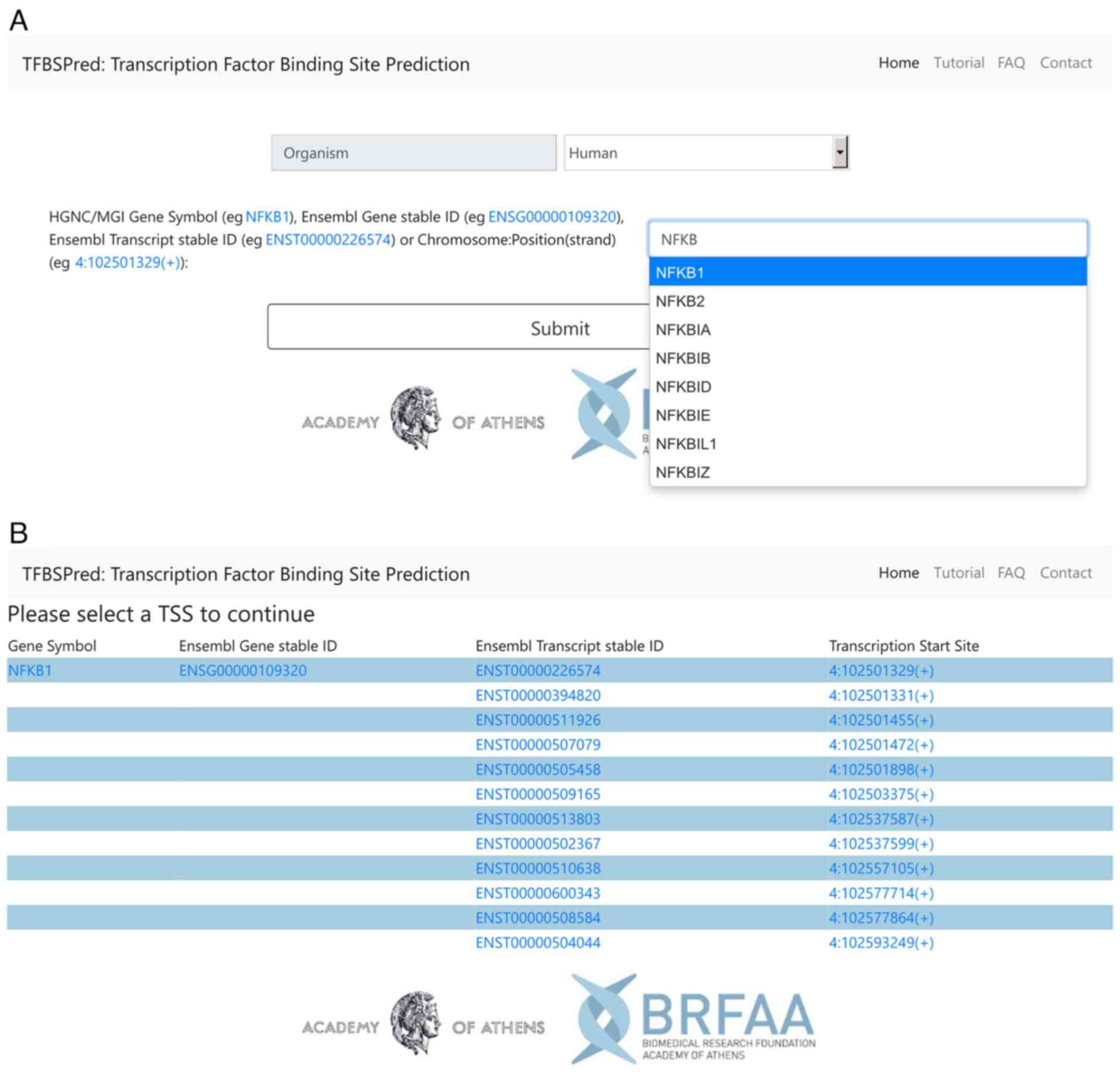
To perform a TFBS search on TFBSPred, the users are guided through a number of online steps, using a web wizard. As input to TFBSPred, the users initially select an organism between human or mouse on the main page (Fig. 1A) and consequently, they type an autocompleted HGNC or MGI gene symbol, respectively, or an Ensembl Gene or Transcript stable ID, or an exact hg38 or mm10 genomic location in the form of Chromosome: Location(Strand), e.g., 4:102501329(+). In the case a gene symbol or Ensembl Gene ID is provided, the users are redirected to the TSS selection page of the gene of interest (Fig. 1B). The TSS selection page provides links to the TSSs of the input gene and to related Ensembl pages. If an Ensembl Transcript ID is used as input, a single link to a TSS will appear on the TSS selection page. By clicking on one of the links to a TSS, the users proceed to the cell type filtering page. The users can directly reach this page from the main page, if they use an exact genomic coordinate input, to perform a TFBS search on a region containing a specific genomic location, instead of a region of a gene promoter.
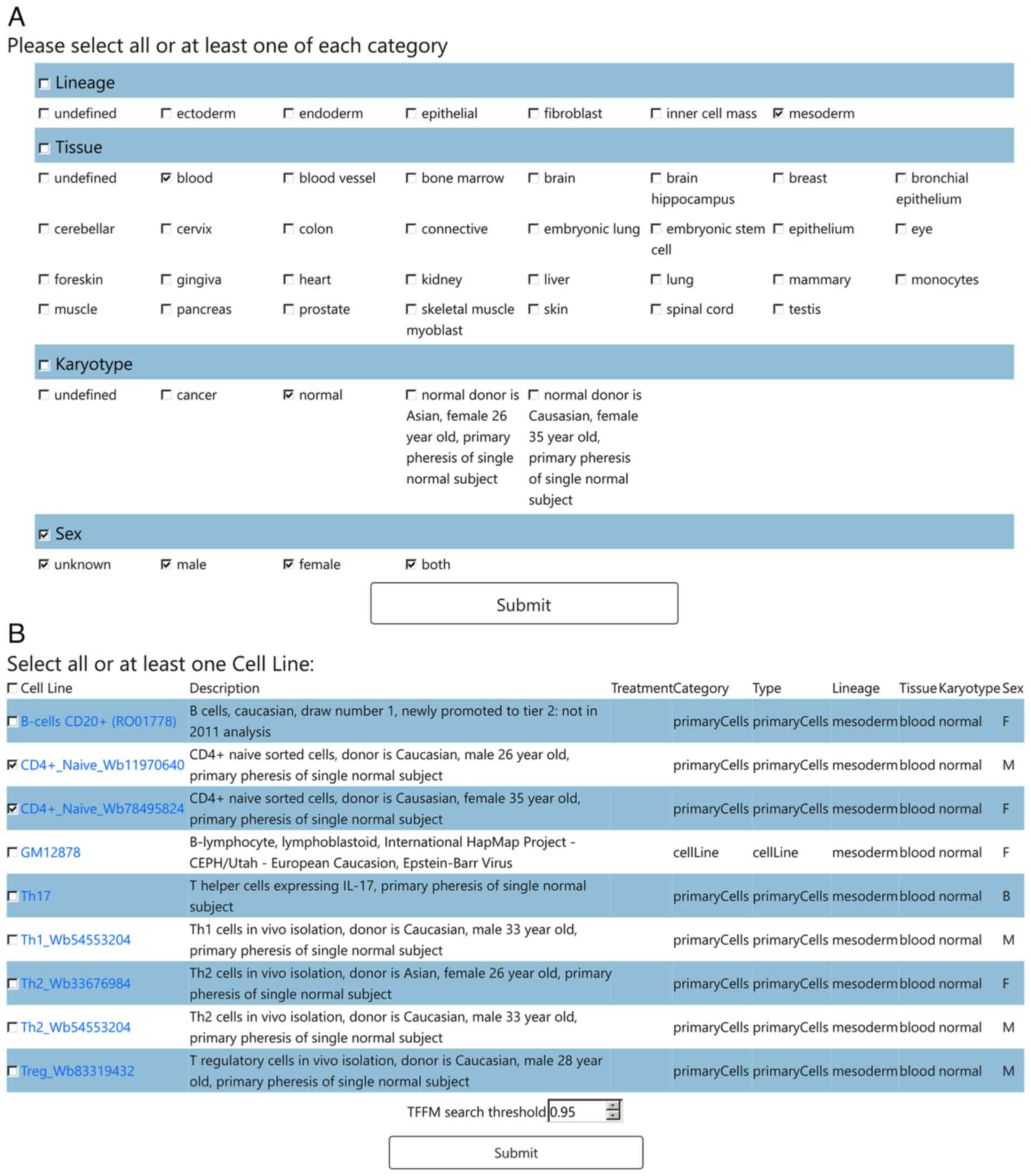
On the cell type filtering page (Fig. 2A), the users filter the cell types whose open chromatin region will be used for analysis, by checking at least one of the boxes of each of four categories: Lineage, Tissue, Sex and Karyotype. By submitting these selections, the users proceed to the cell line selection page (Fig. 2B), where only filtered cell lines are displayed, along with relevant details from Encode. The users check at least one of the boxes of the cell lines. Alternatively, if the users wish to identify all TFs which bind to the promoter region of their gene of interest, irrespective of the tissue context, they need to check all boxes of the cell type filtering and cell line selection pages. To conduct TFFM searches, a TFFM threshold must be specified at the bottom of the cell line selection page. The threshold value ranges from 0.60 to 1, and the higher the number, the stricter the TFBS search is. By submitting the selected cell lines and cut-off value, the TFBS prediction page appears, after a few minutes. A conserved open chromatin region whose extent depends on the cell lines selected, is depicted as a pairwise alignment between human and mouse, with the top sequence belonging to the organism that was initially selected (Fig. 3A). The pairwise alignment follows the ‘pair’ format (http://emboss.sourceforge.net/docs/themes/AlignFormats.html#pair) and can also be downloaded in FASTA format, to be used as input for downstream analyses. The TSS or specified genomic location is marked with an arrow highlighted red, indicating its orientation of transcription. The predicted TFBSs are displayed above their corresponding location in each pairwise alignment with consecutive arrows indicating their binding orientation. Detailed TFBS prediction results can be shown, located below (Fig. 3B). The predicted TFs are sorted alphabetically by their gene symbol, which links to its corresponding JASPAR entry. Each TF contains one or more discovered binding sites presented as a human and mouse pairwise alignment in ‘pair’ format, indicating the actual genomic coordinates of each binding site.
TFFM search
TFBSPred TFBS prediction is based on TFFM searches which are executed in the webtool backend, after the users have submitted their data. To this end, the TFFM framework (22), as well as its prerequisites, the general hidden Markov model (GHMM) (42) and Biopython (43) libraries, were installed.
To define the extent of the open chromatin region of the genome of the organism of interest, which includes the selected TSS or specific genomic location, the borders of the open chromatin region broad peaks of all selected cell lines, which contain the specified genomic coordinate are retrieved from the database and merged together using BEDTools (44), defining the borders of a genomic region which represents the union of all open chromatin regions. The borders of the conserved genomic region which contains the specified genomic coordinate are also retrieved. The conserved open chromatin region is the cross section of the merged open chromatin region and the conserved region, and its borders are calculated using BEDTools. From the pairwise alignment of the conserved open chromatin sequence, the corresponding sequence of the other organism is determined. Both the human and mouse genomic sequences are then searched upon with all TFFMs available, using an in-house python script which outputs the matches above the user designated cut-off value. TFBSPred parses all search results and only displays as pairwise alignments the predicted TFBSs, which are conserved between the human and mouse sequences.
Benchmarking TFBS prediction webtools
TFBS predictions on various human genes were performed using TFBSPred and other webtools, mostly using the default settings. Gene names were submitted to TFBSPred. FASTA-formatted gene regulatory sequences were submitted to AliBaba2.1(45), TFBIND (46), SITECON (47), PROMO (48), MATCH (13) and STAMP (49). Both human and their corresponding mouse sequences were submitted to ConSite (50), FOOTER (51) and rVista 2.0(52). To execute rVista 2.0, the integrated zPicture (53) alignment program was used to align the two sequences. Gene names were used as input and both human and mouse gene promoter sequences were selected, along with JASPAR Core Matrices with all available TFs, in LASAGNA-Search 2.0(54). Gene names were used as input, the exploration function was executed followed by the visualisation function using the discovered TFs, in ConTra v3(55). Finally, the motif features discovered in the regulatory regions of genes in Ensembl regulatory build (56) were also identified.
Results
Interferon beta 1 (IFNB1) enhanceosome
To assess the predictive capabilities of TFBSPred and other TFBS prediction webtools, the IFNB1 enhanceosome (57) was used. The enhanceosome is an ~50 bp enhancer sequence of the IFNB1 gene and requires the coordinate activation and DNA binding of the TFs ATF2/Jun, IRF3 and IRF7 and NF-κB (57,58), effectively dividing the enhanceosome into four positive regulatory domains (PRDI to PRDIV). Τhe exact binding sites of each of these TFs have been experimentally confirmed (57,59). PRDM1 has also been shown to bind specifically to the PRDI domain of IFNB1 enhanceosome, with a negative effect on transcription (60), while additionally playing a role in recruiting co-repressor complexes required to silence gene expression (61). With the default 0.90 TFFM threshold, TFBSPred discovered PRDM1, NF-κB (RELA, RELB and NFKB1), IRF1, SPIB and TEAD2 TFBSs in the enhanceosome region. ATF1 was also discovered among other TFs, when the threshold was decreased to 0.62 (Fig. 4); however, 26 false-positive TFBSs were also predicted (data not shown, as the default cut-off value is not 0.62). The ConTra v3 exploration function predicted only two TFs, NF-κB and PRDM1. AliBaba2.1 discovered five total TFBSs, including IRF8 and ATF2 sites but not NF-κB. LASAGNA-Search 2.0 discovered NF-κB and STAT1, MATCH discovered NF-κB, IRF1 and IKZF1, and the single sequence option of ConSite discovered NF-κB, TEAD1, IRF1 and IRF2. The pairwise alignment option of ConSite only discovered the IRF2 site. PROMO, TFBIND, rVista2.0 and SITECON discovered a large amount of TFBSs. PROMO found TFBSs for all verified TFs; however, it displayed 23 false-positive TFBSs. TFBIND discovered IRF-family TFs, NF-κB and JUN, with 10 false-positive sites. rVista2.0 discovered NF-κB and IRF-family TFs (including IRF7), also having 10 false-positives, and SITECON only found IRF-family TFs, while having 29 false-positive TFBSs. Although STAMP predicted less TFBSs than those of the four previous tools, it only found NF-κB from the verified TFs, presenting 7 false positive sites. Finally, FOOTER did not succeed in predicting any TFBSs. Although the majority of the experimentally verified TFBSs (including NFKB1, NFKB2, IRF3, IRF7, JUN, ATF7 and PRDM1) were computationally predicted by the Ensembl Regulatory Build of IFNB1 (ENSR00001147395), 53 false-positive TFBSs were also found (https://www.michalopoulos.net/tfbspred/erb.html). Nevertheless, none of the putative TFBSs was marked as ‘experimentally verified’ in the Ensembl genome browser (all TFBSs are coloured in grey), indicating that none of the TFBS hits was supported by ChIP-Seq peak evidence.
FGA gene promoter
In its basal state, Fibrinogen alpha chain (FGA) is regulated by HNF1 (-59 to -47 nucleotides from the TSS) and CEBP (-142 to -134 nucleotides from the TSS) family proteins. In an acute state, STAT3 additionally binds to the IL-6RE region, upstream of FGA TSS (62). To examine the tissue-specificity of TFBSPred, two analyses on FGA were conducted: In the first, only endodermal normal cell lines were used, while in the second analysis, only the HepG2 hepatoblastoma cell line was selected. In both cases, the FGA gene was used as input. In the first analysis, TFBSPred discovered CEBP-family factors (CEBPE, CEBPA, CEBPD), HNF1 (HNF1A, HNF1B) and VDR. In the second analysis, a larger conserved open chromatin region was displayed, due to cancer cells having an abnormal regulation pattern. Multiple TFs of the STAT (STAT3, STAT4, STAT5A, STAT5B) and TEAD (TEAD1, TEAD2, TEAD4) families, as well as Bcl6, were additionally discovered further upstream of the TSS (Fig. 5).
Four other webtools that performed best for IFNB1 (single sequence ConSite, rVista2.0, PROMO and ConTra v3), were also used to predict TFBSs for FGA (Fig. 5). The FASTA-formatted human sequence which corresponds to the conserved acute-state open chromatin area upstream to the FGA TSS, as calculated by TFBSPred (345 nucleotides length), was used as input. ConSite predicted only the HNF1 site with 35 false-positives. While rVista2.0 and PROMO all predicted the experimentally verified TFs (HNF1, CEBP and STAT3), they also found 30 and 80 false-positive TFBSs, respectively. ConTra v3 discovered STAT-family TFs and HNF1b, although the latter was not found on the Homo sapiens sequence, but rather in other organisms. TFBIND was also tested as a candidate webtool; however, it was not included since it predicted >400 binding sites (Table SIII).
C-X-C motif chemokine ligand 10 (CXCL10) promoter
CXCL10 is a chemokine that is secreted during the immune response. STAT1, NF-κB, AP1 and heat shock factors bind to an ~230 bp region upstream from the TSS (63). Additionally, IRF3 binds to CXCL10 promoter during hepatitis C infection (64). The CXCL10 gene was used as input to TFBSPred along with all the available cell-lines and default TFFM settings. TFBSPred revealed several factors binding upstream of the gene promoter, including members of the NF-κB (NFKB1, NFKB2 and RELA) family, all the factors forming the NFY complex (NFYA, NFYB and NFYC), as well as IRF1, IRF8, MAFG and STAT2. ConTra v3 exploration analysis discovered only NF-κB family-related TFs (Fig. 6).
Discussion
The most popular approach for TFBS prediction, PWM-based search, considers that the nucleotides of each position exhibit independent participation in the DNA-protein interactions. ConTra v3(55), a widely used webtool, employs PWM-based technologies which use JASPAR and TransFac PWMs to search a given genomic region for potential TFBSs. Specifically, it uses FIMO for its exploration function and MATCH for its visualisation function. The majority of other webtools, such as TFBIND, PROMO and AliBaba2, use their own TFBS prediction algorithms, all of which are based on PWM profiles. The Ensembl Regulatory Build is a collection of regulatory features (denoted as ENSR) across the whole genome. PWMs based on TF motifs which were discovered through SELEX, were matched upon those regulatory regions (34), using MOODS (65), a PWM matching program. However, in the case of the IFNB1 enhanceosome, the loose parameters used for the execution of MOODS in Ensembl Regulatory Build, resulted in low specificity, as >50 pseudo-positive TFBSs were predicted. TFFMs, as a complete HMM-based approach, were designed to address the confounding properties of nucleotide composition, interpositional sequence dependence and variable lengths observed in the recently emerging extensive experimental data. They have been shown to perform more effectively than the majority of PWM-based models (22), while also being publicly available. TFBSPred is the only web based TFBS prediction tool which, thus far, employs the HMM-based TFFM algorithm. In the case of IFNB1, TFBSPred predicted an IRF1 site which coincided with the experimentally verified IRF3 and IRF7 sites in PRDI (57), as well as PRDM1, which was also discovered in the same coordinates. In addition, it should be noted that TFBSPred cannot predict the IRF3 and IRF7 binding sites, as there is no TFFM model for these. The TFBSPred search is connected to the available TFFM profiles from JASPAR. As JASPAR is a continuously evolving project, TFBSPred will incorporate future TFFMs, as well as updates to already existing models. Finally, ATF2/c-Jun leucine zipper heterodimer, does not bind tightly, as the c-Jun half-site is not fully complementary to c-Jun binding and DNA is bent (57). Moreover, the majority of TFFMs for dimer TFs are created for homodimers, rather than heterodimers. Thus, model-based prediction algorithms could not predict this heterodimeric factor. Thus, TFBSPred only predicted ATF1 when the cut-off was lowered, indicating that it is possible to predict difficult TFBS cases, when the TFFM threshold is sufficiently decreased, although this should be used with caution, as it may introduce false-positives.
To reduce the false discovery rate, ConTra v3 takes advantage of the evolutionary conservation of regulatory elements across various related species by expanding the PWM matching on a multiple genome sequence alignment and subsequently visualising the results of the TFBS predictions. However, it is up to the users themselves to evaluate the importance of the conservation of each predicted TFBS among the multitude of species presented. Other webtools, such as rVista2.0 and ConSite, require the user to input two homologous genomic sequences to search for conserved TFBSs by creating a pairwise alignment where conserved TFBSs are automatically determined and displayed. Similar to rVista2.0 and ConSite, TFBSPred searches for conserved TFBSs in a pairwise alignment. However, as opposed to those tools, TFBSPred uses high-quality pairwise alignments from Ensembl Compara, which cover ~1/3 of each genome, while identifying homologous regions between two genomic species is a complex task, which is not easily performed by the average wet lab experimentalists. Another difference is that TFBSPred only compares human and mouse sequences. Although the ability to select between any combination of species and set multiple parameters to search for TFBSs renders rVista2.0 and ConSite more versatile, it markedly increases usage complexity. FOOTER requires the user to input a human and a mouse or rat genomic sequence, and predicts conserved TFBSs between the two; however, its default parameters are not adequate for optimal results. TFBPred requires only one human or mouse genomic sequence as input, and automatically identifies its homologous region of the other species: Mus musculus and Homo sapiens are extensively studied model organisms which belong to the orders of Rodentia and Primates, respectively. These orders which belong to the mammalian superorder of Euarchontoglires, split 85-97 million years ago (66), sharing a high degree of genomic sequence conservation. Thus, predicted TFBSs which are conserved between human and mouse are likely true positives. Searching Compara-aligned human and mouse sequence pairs and automatically discarding TFBSs which are not highly conserved, offers the best possible TFBS results, without any further user participation.
The boundaries of regulatory regions (promoters, enhancers and insulators, as well as TF motifs in open chromatin regions) were defined in Ensembl regulatory build, through a variety of genome-wide experimental data from multiple epigenomic consortia. Predicted TFBSs which are verified by ChIP-Seq can be displayed in the Ensembl genome browser tracks with additional information indicating cell-line/tissue specificity. Although this is an extensive assortment of TFBSs, in the case of the IFNB1 enhanceosome, experimental verification has a low sensitivity, as no experimentally verified TFBSs were identified. As opposed to Ensembl regulatory build, ConTra v3 is unable to define on its own the extent of regulatory regions and has no cell-line/tissue specificity. ConTra v3 requires a user-specified region length to search for TFBSs, with the arbitrary default value being 500 bps. In addition, the majority of webtools perform TFBS prediction solely on a FASTA-formatted genomic sequence the user selects. On the other hand, TFBSPred automatically identifies the boundaries of cell-line/tissue specific regions of open-chromatin which cover around 3-4% of each genome, based on DH data of the selected cell lines, and then searches for conserved TFBSs. Consequently, by selecting certain tissues and cell types, TFBSPred can discover potential cell-type specific TFBSs, as DH areas indicate open chromatin regions that contain regulatory elements of active genes. To examine the basal state of FGA, non-cancer cell lines were used for a TFBSPred analysis: TFBSs for HNF1 and CEBP-family factors were predicted, in accordance with experimentally confirmed sites. To study the acute state of FGA, only Hep2G cancer cell line was used as input for a TFBSPred analysis: Not only was a larger conserved open chromatin region produced, but also STAT family factors were predicted in this expanded open chromatin sequence of the acute state, including experimentally confirmed STAT3. This displays the potential for TFBSPred for tissue-specific differential TFBS predictions, as an experimentally characterised TFBS (STAT3) is predicted in the acute state (Hep2G cancer cell line) and not in the basal state (non-cancer cell lines). R-based BinDNase (67), also incorporates DH data to increase PWM accuracy and has demonstrated the validity of this approach.
ConTra v3 has a long execution period, particularly if a number of PWMs are selected in a visualization analysis. On the contrary, one of the main objectives of TFBSPred was the provision of a rapid and user-friendly interface specifically catering for wet lab biologists. Throughout the website, user interaction is minimal, as the majority of the input fields and selections are provided through TFBSPred database. The TFBSPred web wizard begins with the selection of the TSS of a gene or a chromosomal location in either human or mouse, followed by the filtering and selection of cell types. Finally, the maximum execution time does not exceed a couple of minutes.
To summarise, TFBSPred is a comprehensive and easy-to-use TFBS prediction webtool, based on open chromatin patterns, genomic conservation among human and mouse and HMM-based searches. The present study demonstrated that the predictions of TFBSPred were in accordance with both universal and tissue-specific experimentally verified TFBSs and that in a number of cases, it outperformed existing PWM-based webtools of similar function. TFBSPred is expected to further improve as the TFFM quality and quantity increases. TFBSPred may thus be a useful addition to the experimental biologist community as it provides a working hypothesis that can be experimentally verified.
Supplementary Material
Total open-chromatin region number, average length, standard deviation of the length and percentage of total genomic coverage of each cell line replicate of the DH data for humans and mice.
Total conserved region number, average length, standard deviation of the length and percentage of total genomic coverage, of the pairwise alignment data for humans and mice.
TFBS prediction results of TFBIND, using FGA FASTA formatted sequence as input.
Acknowledgements
The authors wish to acknowledge that the webtool host server is provided by GRNET.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
IM conceived the study. PGB and DAS were also involved in the conception of the study. DH data were downloaded and processed by CN. Multiple alignment between Homo sapiens and Mus musculus data were downloaded and processed by KS. VLZ downloaded the TFFM framework and TFFM profiles, and developed the database and Graphical User Interface (GUI). IK assisted in the website front end development. GAL performed the webtool benchmarking. VLZ and IM confirm the authenticity of all raw data. The manuscript was written by VLZ and IM. All authors contributed to the revision of the work and, have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Wittkopp PJ and Kalay G: Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 13:59–69. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Zawel L and Reinberg D: Initiation of transcription by RNA polymerase II: A multi-step process. Prog Nucleic Acid Res Mol Biol. 44:67–108. 1993.PubMed/NCBI View Article : Google Scholar | |
|
Liu X, Bushnell DA and Kornberg RD: RNA polymerase II transcription: Structure and mechanism. Biochim Biophys Acta. 1829:2–8. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Haberle V and Stark A: Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 19:621–637. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT: The human transcription factors. Cell. 175:598–599. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Lee TI and Young RA: Transcriptional regulation and its misregulation in disease. Cell. 152:1237–1251. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Singh H, Khan AA and Dinner AR: Gene regulatory networks in the immune system. Trends Immunol. 35:211–218. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Narlikar L and Hartemink AJ: Sequence features of DNA binding sites reveal structural class of associated transcription factor. Bioinformatics. 22:157–163. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Stormo GD: Modeling the specificity of protein-DNA interactions. Quant Biol. 1:115–130. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Staden R: Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 12:505–519. 1984.PubMed/NCBI View Article : Google Scholar | |
|
Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al: TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34 (Database Issue):D108–D110. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al: JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48:D87–D92. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV and Wingender E: MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31:3576–3579. 2003.PubMed/NCBI View Article : Google Scholar | |
|
Grant CE, Bailey TL and Noble WS: FIMO: Scanning for occurrences of a given motif. Bioinformatics. 27:1017–1018. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Zhao Y, Ruan S, Pandey M and Stormo GD: Improved models for transcription factor binding site identification using nonindependent interactions. Genetics. 191:781–790. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Khamis AM, Motwalli O, Oliva R, Jankovic BR, Medvedeva YA, Ashoor H, Essack M, Gao X and Bajic VB: A novel method for improved accuracy of transcription factor binding site prediction. Nucleic Acids Res. 46(e72)2018.PubMed/NCBI View Article : Google Scholar | |
|
Toivonen J, Kivioja T, Jolma A, Yin Y, Taipale J and Ukkonen E: Modular discovery of monomeric and dimeric transcription factor binding motifs for large data sets. Nucleic Acids Res. 46(e44)2018.PubMed/NCBI View Article : Google Scholar | |
|
Kulakovskiy I, Levitsky V, Oshchepkov D, Bryzgalov L, Vorontsov I and Makeev V: From binding motifs in ChIP-Seq data to improved models of transcription factor binding sites. J Bioinform Comput Biol. 11(1340004)2013.PubMed/NCBI View Article : Google Scholar | |
|
Rabiner LR: A tutorial on hidden Markov models and selected applications in speech recognition. Proceedings IEEE. 77:257–286. 1989. | |
|
Xu D, Liu HJ and Wang YF: BSS-HMM3s: An improved HMM method for identifying transcription factor binding sites. DNA Seq. 16:403–411. 2005.PubMed/NCBI View Article : Google Scholar | |
|
Wu J and Xie J: Hidden Markov model and its applications in motif findings. Methods Mol Biol. 620:405–416. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Mathelier A and Wasserman WW: The next generation of transcription factor binding site prediction. PLoS Comput Biol. 9(e1003214)2013.PubMed/NCBI View Article : Google Scholar | |
|
Schneider TD and Stephens RM: Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100. 1990.PubMed/NCBI View Article : Google Scholar | |
|
Frazer KA, Elnitski L, Church DM, Dubchak I and Hardison RC: Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13:1–12. 2003.PubMed/NCBI View Article : Google Scholar | |
|
Emes RD, Goodstadt L, Winter EE and Ponting CP: Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum Mol Genet. 12:701–709. 2003.PubMed/NCBI View Article : Google Scholar | |
|
Dolan ME, Baldarelli RM, Bello SM, Ni L, McAndrews MS, Bult CJ, Kadin JA, Richardson JE, Ringwald M, Eppig JT and Blake JA: Orthology for comparative genomics in the mouse genome database. Mamm Genome. 26:305–313. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Elgin SC: DNAase I-hypersensitive sites of chromatin. Cell. 27:413–415. 1981.PubMed/NCBI View Article : Google Scholar | |
|
Pipkin ME and Lichtenheld MG: A reliable method to display authentic DNase I hypersensitive sites at long-ranges in single-copy genes from large genomes. Nucleic Acids Res. 34(e34)2006.PubMed/NCBI View Article : Google Scholar | |
|
Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS and Crawford GE: High-resolution mapping and characterization of open chromatin across the genome. Cell. 132:311–322. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, et al: Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci USA. 101:16837–16842. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al: The accessible chromatin landscape of the human genome. Nature. 489:75–82. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Tweedie S, Braschi B, Gray K, Jones TEM, Seal RL, Yates B and Bruford EA: Genenames.org. The HGNC and VGNC resources in 2021. Nucleic Acids Res. 49:D939–D946. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Smith CL, Blake JA, Kadin JA, Richardson JE and Bult CJ: Mouse Genome Database Group = Mouse Genome Database (MGD)-2018: Knowledgebase for the laboratory mouse. Nucleic Acids Res. 46:D836–D842. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, et al: Ensembl 2019. Nucleic Acids Res. 47:D745–D751. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al: Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxford). 2011(bar030)2011.PubMed/NCBI View Article : Google Scholar | |
|
John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL and Stamatoyannopoulos JA: Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 43:264–268. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al: The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46:D762–D769. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al: The UCSC Genome Browser database: Update 2006. Nucleic Acids Res. 34 (Database Issue):D590–D598. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al: ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 41 (Database Issue):D56–D63. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SM, Amode R, Brent S, et al: Ensembl comparative genomics resources. Database (Oxford). 2016(bav096)2016.PubMed/NCBI View Article : Google Scholar | |
|
Harris RS: Improved pairwise alignment of genomic DNA (unpublished PhD thesis). The Pennsylvania State University, 2007. | |
|
Schliepm A, Georgi B, Rungsarityotin W, Costa IG and Schönhuth A: The general hidden markov model library: Analyzing systems with unobservable states. In: Forschung und wissenschaftliches Rechnen: Beiträge zum Heinz-Billing-Preis. 2004, Gesellschaft f ür wissenschaftliche Datenverarbeitung. Kremer K and Macho V (eds) Gesellschaft für wissenschaftliche Datenverarbeitung mbH Göttingen, Göttingen, pp121-136, 2005. | |
|
Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B and de Hoon MJ: Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25:1422–1423. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Quinlan AR: BEDTools: The swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 47:11.12.1–34. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Grabe N: AliBaba2: Context specific identification of transcription factor binding sites. In Silico Biol. 2 (Suppl):S1–15. 2002.PubMed/NCBI | |
|
Tsunoda T and Takagi T: Estimating transcription factor bindability on DNA. Bioinformatics. 15:622–630. 1999.PubMed/NCBI View Article : Google Scholar | |
|
Oshchepkov DY, Vityaev EE, Grigorovich DA, Ignatieva EV and Khlebodarova TM: SITECON: A tool for detecting conservative conformational and physicochemical properties in transcription factor binding site alignments and for site recognition. Nucleic Acids Res. 32:W208–W212. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J and Alba MM: PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 18:333–334. 2002.PubMed/NCBI View Article : Google Scholar | |
|
Mahony S and Benos PV: STAMP: A web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35:W253–W258. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Sandelin A, Wasserman WW and Lenhard B: ConSite: Web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 32:W249–W252. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Benos PV, Corcoran DL and Feingold E: Web-based identification of evolutionary conserved DNA cis-regulatory elements. Methods Mol Biol. 395:425–436. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Loots GG and Ovcharenko I: rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217–W221. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Ovcharenko I, Loots GG, Hardison RC, Miller W and Stubbs L: zPicture: Dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14:472–477. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Lee C and Huang CH: LASAGNA-Search 2.0: Integrated transcription factor binding site search and visualization in a browser. Bioinformatics. 30:1923–1925. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Kreft L, Soete A, Hulpiau P, Botzki A, Saeys Y and De Bleser P: ConTra v3: A tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 45:W490–W494. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Zerbino DR, Wilder SP, Johnson N, Juettemann T and Flicek PR: The ensembl regulatory build. Genome Biol. 16(56)2015.PubMed/NCBI View Article : Google Scholar | |
|
Panne D: The enhanceosome. Curr Opin Struct Biol. 18:236–242. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Merika M and Thanos D: Enhanceosomes. Curr Opin Genet Dev. 11:205–208. 2001.PubMed/NCBI View Article : Google Scholar | |
|
Panne D, Maniatis T and Harrison SC: An atomic model of the interferon-beta enhanceosome. Cell. 129:1111–1123. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Keller AD and Maniatis T: Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5:868–879. 1991.PubMed/NCBI View Article : Google Scholar | |
|
Elias S, Robertson EJ, Bikoff EK and Mould AW: Blimp-1/PRDM1 is a critical regulator of type III Interferon responses in mammary epithelial cells. Sci Rep. 8(237)2018.PubMed/NCBI View Article : Google Scholar | |
|
Fish RJ and Neerman-Arbez M: Fibrinogen gene regulation. Thromb Haemost. 108:419–426. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Vazirinejad R, Ahmadi Z, Kazemi Arababadi M, Hassanshahi G and Kennedy D: The biological functions, structure and sources of CXCL10 and its outstanding part in the pathophysiology of multiple sclerosis. Neuroimmunomodulation. 21:322–330. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Brownell J, Bruckner J, Wagoner J, Thomas E, Loo YM, Gale M Jr, Liang TJ and Polyak SJ: Direct, interferon-independent activation of the CXCL10 promoter by NF-κB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol. 88:1582–1590. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Korhonen JH, Palin K, Taipale J and Ukkonen E: Fast motif matching revisited: High-order PWMs, SNPs and indels. Bioinformatics. 33:514–521. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Kumar S, Stecher G, Suleski M and Hedges SB: TimeTree: A resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Kahara J and Lahdesmaki H: BinDNase: A discriminatory approach for transcription factor binding prediction using DNase I hypersensitivity data. Bioinformatics. 31:2852–2859. 2015.PubMed/NCBI View Article : Google Scholar |