Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum
- Authors:
- Published online on: July 24, 2017 https://doi.org/10.3892/ijmm.2017.3080
- Pages: 834-844
-
Copyright: © Tang et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Extracellular vesicles (EVs) are small particles (30–1000 nm) secreted by various types of cells; they are enclosed by a phospholipid bilayer and contain DNA, RNA and protein (1). EVs can be released into cell culture medium (CCM), and they are also found abundantly and naturally in body fluids. They may therefore serve as biomarkers for the development of superior, sensitive and minimally invasive diagnostic alternatives in 'precision medicine' (2,3). Due to the small size and heterogeneity of vesicles, EV detection and classification is challenging. Different types of vesicles have been identified, such as exosomes, microvesicles, apoptotic bodies, secreted proteins and retrovirus-like vesicles (1,4). In the present study, the widely used term 'exosomes' was used to refer to EVs (exosomes and other types of cell-derived vesicles) in general.
The commonly used protocol for isolation of exosomes is ultracentrifugation (UC); the final step of which is centrifugation at 100,000 × g at least for 70 min to pellet the small vesicles that correspond to exosomes (5). In addition, sucrose density gradients, ultrafiltration (6), high performance liquid chromatography-based protocols (7) and immunoaffinity-capture methods (8), singly or combined with the application of UC, can provide high enrichment and purity of exosomes (9). In recent years, easy-to-use precipitation solutions, such as ExoQuick and Total Exosomes Isolation Reagent (TEI), have been utilized to precipitate particles in liquid. The procedure is convenient and time-saving with no need for expensive equipment or technical challenge (10,11). However, the 'salting out' methods, unable to resolve particle heterogeneity, are not specific for exosomes or other EVs and may easily lead to the isolation of non-exosomal particles (12).
In the downstream analysis of exosomal content, a number of alternative exosomal RNA (exoRNA) extraction methods have been used, including phenol-based techniques (TRIzol) and combined phenol and pure column-based techniques [miRNeasy and HiPure Liquid RNA/miRNA kit (HLR)] (13). Recently, commercial kits [SeraMir™ Exosomes RNA Amplification kit (SeraMir), Total Exosomes RNA and Protein Isolation kit (TER)] have been designed specifically for the isolation of RNA and protein from a single enriched exosome preparation. The exoRNeasy Serum/Plasma kits use a membrane-based affinity binding step to isolate exoRNA directly from serum or plasma (14).
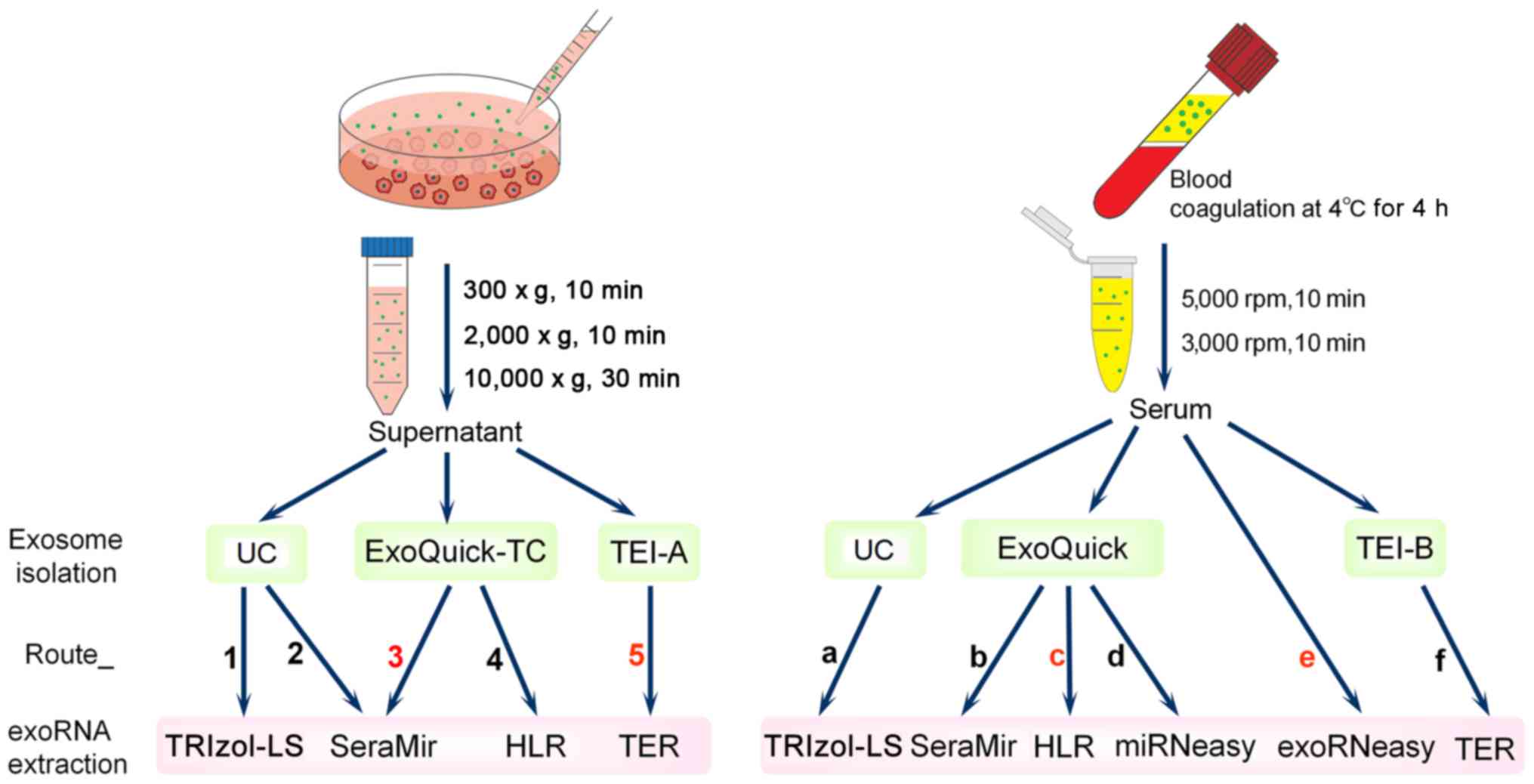
Given the highly attractive research value of exoRNA, the availability of convenient methods to extract the exoRNA with high quality, substantial yield, purity and appropriate size distribution needs to be confirmed. Recently, comparative studies on the impact of isolation methods for either exosomes (15,16) or exoRNA (13,17) on downstream RNA yield, quality and profiles have been conducted. However, in practice, the methods used for extraction of both exosomes and exoRNA should be considered in order to extract exoRNA of high-quality. In addition, a large range of methods including several new commercial kits designed specifically for exoRNA extraction (SeraMir, TEI and exoRNeasy) should be utilized and compared. Therefore, in this study (Fig. 1), nanoparticle tracking analysis (NTA) and protein analysis were used to measure and characterize CCM or serum-derived exosomes isolated using UC and two commercially available kits (ExoQuick and TEI). Subsequently, exoRNA was isolated by a combination of the above methods for exosome isolation and six exoRNA isolation methods (TRIzol-LS, SeraMir, TEI, HLR, miRNeasy and exoRNeasy). Given that there were too many methods for each combination to be tested, we determined the combination patterns (Route_1 to Route_5 for CCM and Route_a to Route_f for serum) (Fig. 1) according to the recommendations of the relevant manufacturers or other published studies (11,17–20). The quantity and quality of exoRNA were determined using NanoDrop and Bioanalyzer 2100. By the use of qPCR and high-throughput sequencing analysis, we provide evidence that different combinations of isolation methods for exosomes and exoRNA can affect exoRNA profiling.
Materials and methods
Study design and participant consent
This study was approved by the Ethics Committee of Nanfang Hospital, Guangzhou, China, and written informed consent was obtained from all participants. The sample collection and treatment were carried out in accordance with the approved guidelines. The experiment was repeated three times using three completely independent sets of samples (three independent CCM or serum samples prepared at different times). Each sample was divided into 100 ml CCM and 500 µl serum for each extraction method. A flowchart of the study design is shown in Fig. 1. The human lung cancer cell line A549 (ATCC, Manassas, VA, USA) was cultured in serum-free RPMI-1640 medium and 2% Exo-FBS™ exosome-depleted fetal bovine serum (System Biosciences, Mountain View, CA, USA) for 48 h and the CCM was collected and centrifuged at 300 × g for 10 min, then at 2,000 × g for 10 min, and finally at 10,000 × g for 30 min to remove dead cells, cell debris and large particles (shedding vesicles and apoptotic bodies). Blood samples were obtained from three healthy donors (mean age, 28 years; gender, one female and two males) at the Department of Laboratory Medicine, Nanfang Hospital, Southern Medical University, Guangzhou, China. Blood in containers without anticoagulant or coagulant was kept at 4°C for 4 h to ensure serum separation. Serum samples were centrifuged at 5,000 rpm for 10 min, and then at 3,000 rpm for 10 min and stored at −80°C before use.
Methods for exosome isolation from CCM or serum included: UC, ExoQuick (System Biosciences) and TEI (Invitrogen, Life Technology, Carlsbad, CA, USA). RNA was extracted from exosomes using four different methods: TRIzol-LS (Ambion, Life Technology, Carlsbad, CA, USA), SeraMir (System Biosciences), TER (Invitrogen), HLR (Magen, Guangzhou, China) and miRNeasy (Qiagen, Hilden, Germany). ExoRNeasy (Qiagen) is able to purify exosomal RNA from serum directly.
Exosome isolation
The UC method was used as previously described (5). The supernatant was ultracentrifuged using a W32Ti rotor (L-80XP; Beckman Coulter, Brea, CA, USA) at 110,000 × g for 70 min to pellet the exosomes. The pellet was washed in phosphate-buffered saline (PBS) to eliminate contaminating proteins, and centrifuged again at 110,000 × g for 70 min. The PBS was removed and the exosomes re-suspended in 100 µl PBS or nuclease-free water. The nanomaterial exosome isolation methods tested comprised four kits (ExoQuick-TC, TEI for CCM, ExoQuick, TEI for serum), which were used according to the manufacturer's instructions. All centrifugation steps were performed at 4°C.
Nanoparticle tracking analysis (NTA)
Vesicle suspensions with concentrations between 1×107/ml and 1×109/ml were examined using a Nanosight NS300 (NanoSight Ltd., Amesbury, UK) equipped with a 405 nm laser to determine the size and quantity of particles isolated. A video of 60-sec duration was taken with a frame rate of 30 frames/sec, and particle movement was analyzed using NTA software (version 2.3; NanoSight Ltd.).
Transmission electron microscopy (TEM)
A 20–40 µl solution of exosomes was placed on a copper mesh and post-negatively stained with 2% phosphotungstic acid solution for 10 min. The sample was then dried for 2 min under incandescent light. The copper mesh was observed and photographed under a transmission electron microscope (H-7650 Hitachi microscope; Hitachi, Tokyo, Japan).
Western blot analysis
The exosome supernatant was denatured in 5X sodium dodecyl sulfonate (SDS) buffer and subjected to western blot analysis (10% SDS-polyacrylamide gel electrophoresis; 50 µg protein/lane) using rabbit polyclonal antibody CD63 (sc-15363) in CD9 (sc-13118; both from Santa Cruz Biotechnology, Inc., Dallas, TX, USA), TSG101 (T5701; Sigma-Aldrich, Dorset, UK) and calnexin (BS1438; Bioworld Technology, St. Louis Park, MN, USA). The proteins were visualized on the Bio-Rad ChemiDoc XRS Imager system (Bio-Rad Laboratories, Berkeley, CA, USA).
ExoRNA isolation and RNA analyses
TRIzol-LS reagent was designed to isolate high-quality total RNA from liquid samples, and 250 µl of exosome solution was lysed in 750 µl TRIzol-LS. Subsequently, 200 µl of chloroform was used for phase separation and 100% isopropanol for RNA precipitation. Finally, RNA was eluted in 30 µl RNase-free water after being washed twice in 75% ethanol. TER, HLR, miRNeasy, SeraMir and exoRNeasy kits were used according to each manufacturer's total RNA isolation procedure.
The RNA concentration was assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The RNA yield and size distribution were analyzed using an Agilent 2100 Bioanalyzer with an RNA 6000 Pico kit (Agilent Technologies, Foster City, CA, USA).
RNA profiling analysis
Aliquots containing 100 ng of three exoRNA samples from CCM isolated by different methods were used for RNA library preparation, following the instructions for the NEBNext Multiplex Small RNA library preparation kit (New England Biolabs, Ipswich, MA, USA). The PCR amplified cDNA construct (from 140–160 bp) was purified using a QIAquick PCR Purification kit (Qiagen). The purified cDNA was directly sequenced using an Illumina MiSeq 2000 platform (Illumina, San Diego, CA, USA). The miRNA contained in serum exosomes was analyzed using an All-in-One™ miRNA First-Strand cDNA synthesis kit and miRNA qPCR kit (GeneCopoeia, Rockville, MD, USA).
Statistical analysis
One-way or two-way analysis of variance (ANOVA) was used to investigate the differences between groups. Differences in paired samples were compared using the two-tailed Student's t-test. Correlations between variables were assessed by Pearson's correlation. Linear regression was applied to determine the linear regression equation. SPSS 15.0 was used for statistical analyses (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to be statistically significant.
Results
Among the three exosome isolation methods (UC, ExoQuick and TEI), UC shows the lowest yield and recovery, but the highest protein purity
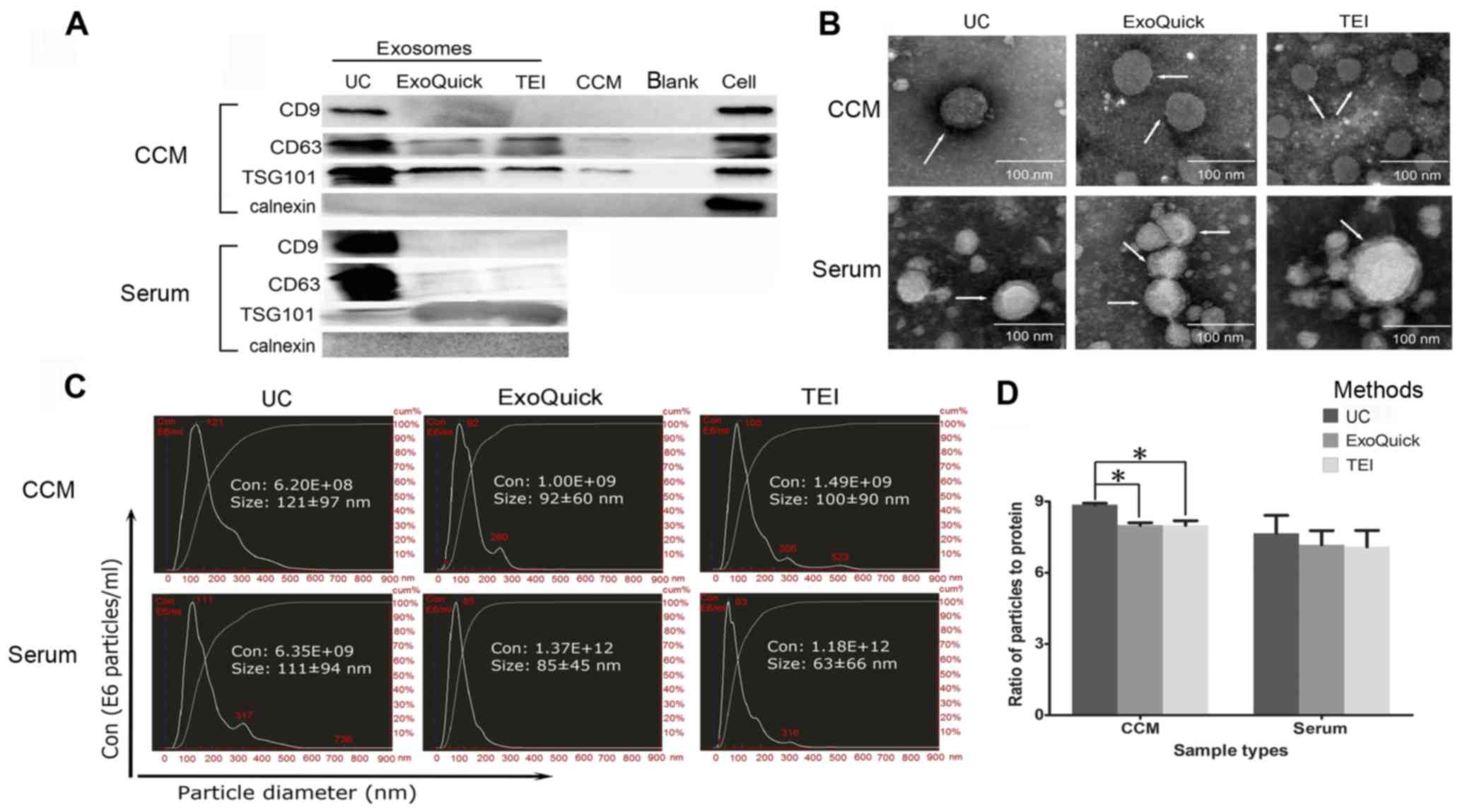
To identify exosomes, the vesicles isolated from A549 CCM and serum were investigated by western blot analysis (Fig. 2A), TEM (Fig. 2B) and NTA (Fig. 2C). The existence of exosomal protein markers (CD9, CD63 or TSG101) and non-existence of non-exosomal markers (calnexin), the lipid bilayer structure, and the size of particles (50–200 nm) were used to demonstrate the presence of exosomes.
The recovery rate of exosomes generated by the three methods was compared using NTA (Table I). Either in CCM or serum, the two commercial kits (ExoQuick and TEI) showed higher exosomal recovery than UC. In CCM, TEI produced a higher yield of exosomes than ExoQuick. However, in serum, no difference was found between the two commercial kits.
Exosomes are commonly quantified using protein concentration and particle numbers. The ratio of particle number to protein concentration has been used to assess co-isolation of protein contaminants and exosome purity (12). Overall, significant differences in this ratio were found among the three exosomes isolation methods (UC, ExoQuick and TEI) in CCM (P=0.005) (Fig. 2D). Exosomes isolated by UC showed a higher ratio than those from ExoQuick (UC vs. ExoQuick: 8.86±0.10 vs. 8.00±0.18, P=0.024) and TEI (UC vs. TEI: 8.86±0.11 vs. 7.99±0.34, P=0.049) in CCM (Fig. 2D), indicating the lowest level of protein contaminants and highest purity. No differences were found between ExoQuick and TEI. Although the differences were not significant (P>0.05), the same trend for UC to have a higher ratio was also observed in serum (UC vs. ExoQuick vs. TEI: 7.66±1.30 vs. 7.17±1.03 vs. 7.09±1.19) (Fig. 2D). Interestingly, the finding that exosomal markers were the most highly enriched in UC exosome preparations (Fig. 2A) also demonstrated a higher purity than that obtained with ExoQuick and TEI.
Upon comparison of exoRNA extraction, Route_3 and Route_e show the highest quantity and recovery among the five extraction methods for CCM and the six methods for serum, respectively
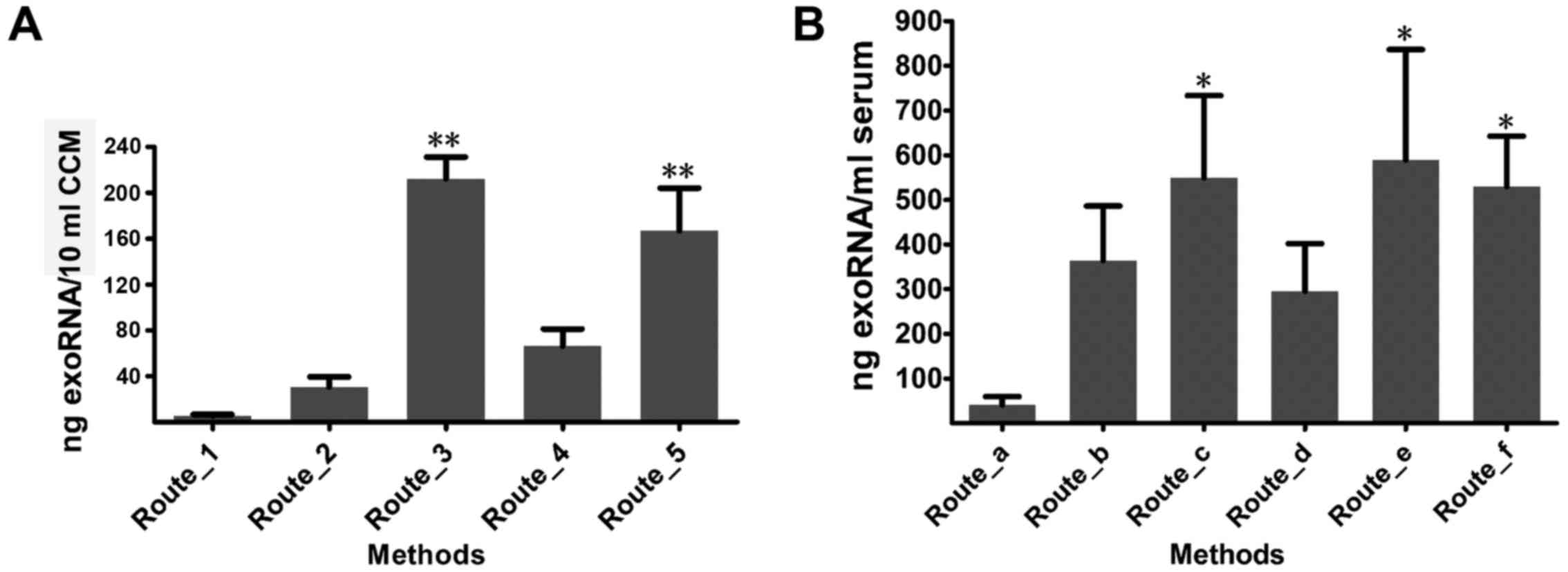
To evaluate the influence of isolation methods on the exoRNA yield of CCM, five combinations of extraction methods for exosomes and exoRNA (Route_1 to Route_5) were compared (Fig. 3A). The experiment was repeated three times using three CCM samples. Among these methods, Route_3 showed the highest extraction efficiency (212.13±19.14 ng RNA/10 ml CCM), and was followed by Route_5 with a concentration of 167.06±37.02 ng RNA/10 ml CCM. In addition, the RNA concentrations obtained using Route_3 and Route_5 were significantly higher than those from the other three routes [Route_1 (5.48±1.25), Route_2 (30.71±8.84) and Route_4 (66.18±15.05)] (P<0.01). Overall, when a high exoRNA yield from CCM is needed, Route_3 and Route_5 are the recommended methods.
With respect to exoRNA isolation from serum, six combinations of methods (Route_a to Route_f) were used. The experiments were repeated three times using samples from three different donors (Fig. 3B) which may explain the variation in exoRNA quality. Overall, Route_e recovered the most exoRNA among all the methods (589.20±247.26 ng RNA/ml serum), and was followed by Route_c (549.00±184.66 ng RNA/ml serum) and Route_f (530.00±112.69 ng RNA/ml serum). Route_c, Route_e and Route_f extracted a higher yield of exoRNA than Route_a (40.45±19.21 ng/ml serum) (P<0.05). Therefore, on the basis of exoRNA yield, Route_e, followed by Route_c and Route_f could be recommended for serum exoRNA isolation.
The exoRNA obtained using Route_4 and Route_5 (for CCM), or Route_e (for serum) shows the highest yield and the most uniform size of small RNA
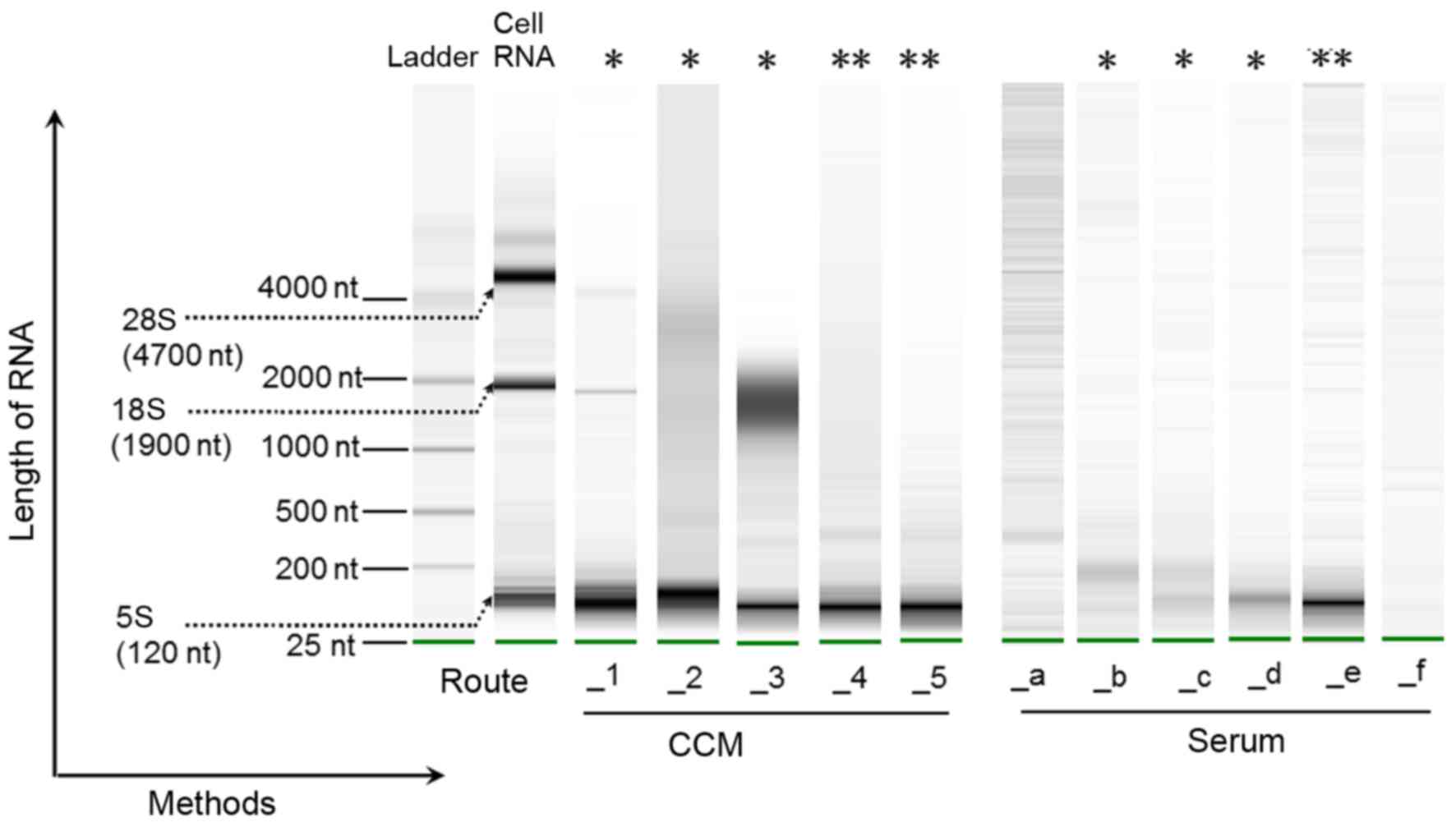
Total exoRNA recovery from CCM and serum was characterized using an Agilent 2100 Bioanalyzer (Fig. 4). It was shown that the majority of exoRNA from CCM was small (<200 nt), but there were variations in RNA quantity and quality among the different methods. Route_1, Route_2 and Route_3 presented not only the main peak around 100 nt but also some longer RNA species including 18S and 28S rRNA. However, Route_4 and Route_5 showed the best quality of small RNA which was free of 18S RNA contamination; therefore, Route_4 and Route_5 may be more suitable for small exoRNA research.
For serum exoRNA, 18S and 28S ribosomal RNA were absent from the samples produced by all methods (Fig. 4). Overall, Route_e (exoRNeasy) resulted in the highest yield and the most uniform size of small RNA, indicating the highest extraction efficiency of small exoRNA from serum among the six combinations of methods (Route_a to Route_f).
Isolation methods influence small RNA constituent ratio, miRNA content and amount
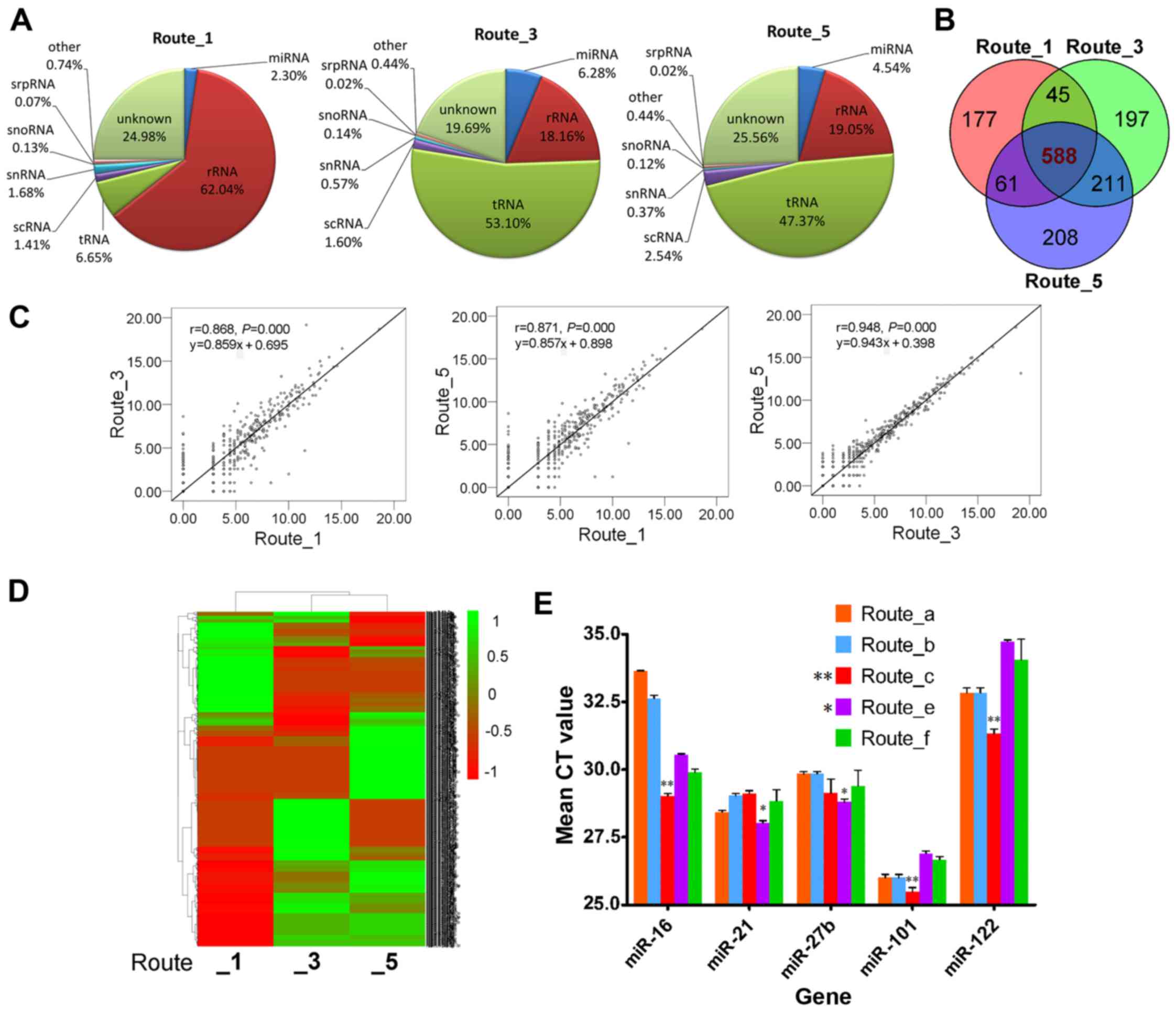
To examine the effect of the isolation methods on the exoRNA profile of CCM, RNA sequencing analysis was performed using three exoRNA samples (100 ng exoRNA/sample) isolated from the same CCM by three commonly used and representative routes: Route_1 isolated by traditional UC and TRIzol-LS methods, Route_3 isolated by kits from the Systems Biosciences (SBI) Company (ExoQuick and SeraMir), and Route_5 isolated by kits from Life Technology (TEI and TER). Among the three samples, we obtained 12.56 million raw reads from Route_1, 9.73 million reads from Route_3 and 12.61 million reads from Route_5. After trimming low-quality reads, 5,994,511 (51.21%) clean reads from Route_1, 8,030,999 (87.80%) from Route_3 and 9,379,868 (83.86%) from Route_5 could be mapped to known RNAs of the human genome. These mappable sequences were annotated to miRNA and other small non-coding RNAs. The percentages of miRNA, rRNA, tRNA and other RNA are shown in a pie chart (Fig. 5A): rRNA or tRNA was the most abundant small RNA in the three samples. Route_1 samples mainly contained rRNA (62.4% of all small RNA), but tRNA was the main RNA in samples from Route_3 (53.1%) and Route_5 (47.37%). In addition, Route_3 (6.28%) yielded a higher percentage of miRNA than Route_1 (2.3%) and Route_5 (4.54%) (Fig. 5A).
We defined detectable miRNAs as those that had at least one sequence per million mappable miRNA reads. Accordingly, we detected 402 known hsa-miRNAs in samples from Route_1, 570 known hsa-miRNAs in samples from Route_3 and 574 known hsa-miRNAs in samples from Route_5. To demonstrate the methodological variations, we investigated whether the miRNAs were unique to or common to the different exoRNA preparation protocols. Among all detectable miRNAs, 588 miRNAs were detected in samples obtained using all three routes (Fig. 5B). In addition to the miRNAs shared by three samples, or at least by two samples, some miRNAs were also identified that were unique to each isolation method. For example, miR-33b-3p, miR-500a-5p and miR-1247-3p were only detected in exoRNA samples isolated by Route_1, while miR-433-3p, miR-338-5p and miR-1262 were only found with Route_3 and miR-219a-5p, miR-30c-1-3p and miR-616-5p only with Route_5.
The 100 most abundant miRNAs made up 95.3–96.9% of the detectable miRNA sequences, among which the top 20 abundant miRNA are shown in Table II. The top 20 abundant miRNAs obtained with the different isolation methods were similar, but methodological variation was also evident in these miRNAs. The rank of each miRNA obtained by the three routes was different. To examine the variation in the miRNA content resulting from methodological variability, a correlation analysis was performed (using log2 transformed values after normalizing reads to per million counts). Overall, correlations were found among the three pairs of samples (P=0.000 for all) (Fig. 5C). The correlation (r) values were 0.868 between Route_1 and Route_3, 0.871 between Route_1 and Route_5, and 0.948 between Route_3 and Route_5 (Fig. 5C), indicating that similar samples were produced by the two commercial kits used in Route_3 and Route_5. However, hierarchical clustering analysis (Fig. 5D) showed that significant differences existed among the methods, although there was higher similarity between Route_3 and Route_5.
Table IIThe 20 most abundant exosome miRNA of three samples isolated by the different methods from CCM. |
Given the limited serum volume, we were unable to extract sufficient RNA for sequencing analysis. Therefore, the recovery of exosomal microRNA from serum was compared by qPCR. To confirm the presence and amount of exosomal microRNA in serum, five microRNAs (miR-16, miR-21, miR-27b, miR-101 and miR-122) earlier reported to be present in the circulatory system or exosomes were analyzed by qPCR (Fig. 5E). Overall, the miRNA showed significant differences (P<0.05) among the different routes. Specifically, the cycle threshold (CT) value of miR-16 was highest for Route_a (33.64±0.03), indicating the lowest copy number in the extracted sample. Route_c resulted in the highest yield of miR-16 (27.05±0.53). In addition, the amounts of miR-101 (25.49±0.27) and miR-122 (31.33±0.27) were also highest for Route_c, indicating that Route_c provided high and stable extraction efficiency for miRNA in serum. Route_e showed changeable profiling, and produced the lowest level of miR-101 (26.91±0.15) and miR-122 (34.73±0.09) but the highest level of miR-21 (28.8±0.19) and miR-27b (28.03±0.13), indicating selectivity of the different miRNAs isolated by this method.
Discussion
In the present study, different traditional methods and commercial kits were tested and compared to define the most suitable isolation protocols for exosomes or exoRNA for use with CCM and serum samples. The results were as follows. i) For exosome isolation, two commercial kits (ExoQuick and TEI) showed higher extraction efficiency than traditional UC, but UC samples had less protein contamination than those from ExoQuick and TEI. ii) Route_3 and Route_5 showed high and stable exoRNA recovery from CCM. iii) Route_e, followed by Route_c and Route_f, showed high exoRNA recovery from serum; iv) high yield and narrow size distribution pattern of small RNA presented in exoRNA isolated by Route_4 and Route_5 from CCM, and Route_e from serum. v) RNA sequencing analysis proved that isolation method can influence the small RNA profile.
As a general rule, at least two different technologies should be used to characterize individual EVs. Therefore, three methods, western blot analysis, TEM and NTA, were used to demonstrate the presence of exosomes in this study. According to the minimal requirements for EV studies suggested by Lötvall et al (21), investigators should report the amount of several (three or more) 'exosome-enriched' and non-EV protein markers in any EV preparation. Calnexin, the marker for non-EV components, was not expressed in any of the exosome samples, indicating the absence of contamination with intracellular proteins during exosome purification. The cytosolic protein (TSG101) and the transmembrane protein (CD63) were present in all exosome preparations. However, the other transmembrane protein (CD9) was found only in exosomes isolated by UC, but not in those obtained using the two kits. As Lötvall et al recommend (21), the composition of EVs should ideally be compared with that of the secreting cells. We found that exosomes isolated by UC had a comparable (CD9) or even greater (CD63 and TSG101) level of enrichment of the EV components than the originating cells. The weak bands were found in CCM, indicating that the supernatant 70 min post ultracentrifugation still contained significant quantities of the remaining EVs (22).
NTA technology can rapidly calculate the total number and the overall size distribution of vesicles. However, a criticism of this technology, which is based on light scattering and Brownian motion, is that it cannot differentiate adequately among synthetic nanoparticles, large protein aggregates and biologic vesicles (23). Although protein quantification is the method most frequently used to estimate the number of exosomes, it may overestimate this number by detecting contaminating proteins that are not associated with exosomes (24). On considering the advantages and disadvantages of the two approaches, a combination of both particle number and protein concentration analysis was used to analyze the efficiency and purity of exosome isolation. Currently, the ratio of particles to protein concentration (particle number/microgram of protein) is accepted as a good indicator of particle purity. Some precipitation protocols produce highest high yield of particles, yet have low ratios of particles to protein, potentially due to co-isolation of contaminants (12,25).
Our analysis of NTA showed that particle numbers obtained using UC were lower than those of the two commercial kits (ExoQuick and TEI), but the results of western blotting showed that samples isolated by UC methods were more enriched for exosome markers. This may be caused by the following: i) biologic vesicles may be damaged by repeated ultracentrifugation steps, which causes the low particle recovery of UC methods; ii) polymeric precipitation (ExoQuick and TEI) shows higher particle recovery, but the lack of a method to separate exosomes and high-density protein aggregates may also result in contamination with free proteins in the exo-pellet samples, especially in serum samples with abundant albumin (26). In addition, the ratio of particles to protein also suggests different degrees of protein contamination in samples from the three methods. The UC method provided the lowest exosome recovery rate, but a higher ratio indicated higher protein purity of exosomes obtained by UC than with ExoQuick and TEI. Therefore, UC is more suitable for proteomic research, which requires higher protein purity of exosomes. Notably, our results are consistent with the general consensus but partially in opposition to the research of Caradec's group (24). They found less albumin contamination and better exosome enrichment in exosomes isolated by ExoQuick rather than by UC. They based this conclusion on the western blot result of LAMP2 but not on the exosome markers (CD9, CD63 and TSG101) used in this study. The relative proportions of different proteins (including exosome markers) seem to vary in the different types of EV (21). Therefore, subsets of EVs isolated by various methods may be different. The amount of blood collected from patients may be limited, and although UC showed higher protein purity of exosomes, it is not always applicable to clinical samples given the large volume of starting material required and the low extraction efficiency of this method. A recent study reported that the qEV column from Izon Science Company which extracted exosomes on the basis of size exclusion chromatography, provided exosomes with both superior purity and higher exosome recovery rate than UC combined with density gradient purification, and therefore this may be an alternative method for proteomic research using CCM or clinical samples (12). Although ExoQuick and TEI samples may be contaminated with proteins, in some exoRNA research, when the analysis may be less influenced by proteins, the two kits may be good choices because of their high extraction efficiency.
In the RNA isolation step, we compared the RNA yield and purity of the different combinations of traditional or commercial extraction methods for exosomes and exoRNA to define the most suitable exoRNA isolation protocol for CCM. Overall, Route_3 showed the highest exoRNA recovery from CCM and Route_1 showed the lowest exoRNA recovery. Because Route_3 uses dispersive electrophoresis strips not only in the small RNA area (<200 nt) but also in the long RNA area (1000–2000 nt), it may be more suitable for total RNA research rather than small RNA research. Although the Route_3 methods (ExoQuick-TC and SeraMir) are often recommended for use in combination, we found that either ExoQuick-TC combined with other RNA isolation methods (Route_4) or SeraMir combined with other exosome isolation methods (Route_2) was also feasible. Despite the fact that the exoRNA recovery is reduced, the long RNA banding weakens or even disappears after either the exosome or the exoRNA extraction method is changed. These changes would be more applicable for small RNA analysis. Intriguingly, Route_5 showed high exoRNA recovery and also small RNA bands with higher quality, indicating superior methods for CCM small exoRNA research.
For serum exoRNA extraction, Route_e (ExoRNeasy) showed high recovery yield and also the most apparent and narrow well-size distribution pattern of small RNA. In addition, researchers can isolate exoRNA by using this kit alone without a specific exosome isolation process. The spin columns have the capacity to handle up to a 4-ml sample volume, enabling use of finite resources such as clinical samples or concentrated CCM. Therefore, exoRNeasy is an improvement, and is a rapid method that could be easily adapted to the clinical laboratory (14). Of note, although the miRNeasy (in Route_d) and exoRNeasy kits came from the same company, exoRNeasy is specifically designed for exoRNA isolation. Our finding that Route_e showed a more obvious band of small RNA than Route_d indicated that exoRNeasy was more suitable for exoRNA isolation than miRNeasy. Moreover, Route_b, Route_c can be used as an alternative method for extraction of exoRNA owing to the high RNA recovery and visible bands in the small RNA area.
In the last step, the RNA cargo of the isolated exosomes was analyzed. High sequencing analysis of CCM exoRNA profiling revealed that the RNA content was similar, but some differences existed among samples obtained using the various isolation methods. Firstly, the primary data showed that more clean reads were found in samples from commercial kits (Route_3 and Route_5) than from the traditional Route_1. In addition, the final sequencing results after mapping the data base revealed that higher proportions of miRNA were found in exoRNA isolated by kit methods than with UC. Therefore, Route_3 and Route_5 are also applicable to the preparation of exoRNA for small RNA sequencing, and may have more advantages for miRNA sequencing. Finally, most miRNAs were common to the three methods and Pearson correlation analysis demonstrated good correlations among them. However, a higher correlation coefficient was found between the two kits (Route_3 and Route_5), and we also discovered more similarity between the two kit methods in constituent ratio analysis of small RNA and cluster analysis of miRNA. These results may be explained by the similar experimental principles used in Route_3 and Route_5.
With regard to the miRNA expression of exosomes in serum, among the three methods tested Route_c had the highest level of detected miRNA, indicating a good and stable extraction efficiency of miRNA. Variations in miRNAs were also found in samples obtained using the different routes. For example, Route_e performed best for miR-21 and miR-27b but worse for miR-101 and miR-122 expression. In contrast, Route_c performed worst for miR-21 but best for miR-101 and miR-122 expression. A conclusion from our experiments is that different isolation methods have different affinity and performance for specific miRNA molecules, which is in good agreement with previous studies (13,15,27).
The major limitation of the study is that each exosome isolation method was grouped together with a particular exoRNA purification procedure, and not with the others. Although this design is helpful in finding the optimum extraction mode, the differences in the initial exosome isolation procedure could affect the results of exoRNA isolation. Considering that some researchers have decided on the exosome isolation method to be used in their laboratories, they may be more concerned with the efficiency of exoRNA extraction methods. Therefore, in further studies, the same initial exosomes should be used separately to compare each exoRNA isolation method.
A conclusion from this study is that different isolation methods may account for the concentration, purity and size discrepancies of exosomes or exoRNA, therefore it is important to maintain consistency and use one isolation method for each research application. This study also showed advantages and disadvantages of each method and their application to different types of research (Table III), therefore providing a reference for use when choosing an isolation method according to research design.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (grant no. 81371901), the Science and Technology Program of Guangzhou (grant no. 1563000220).
References
|
van der Pol E, Böing AN, Harrison P, Sturk A and Nieuwland R: Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 64:676–705. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B, Inal JM and Zheng L: Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 4:275222015. View Article : Google Scholar : PubMed/NCBI | |
|
Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al: Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Cocucci E, Racchetti G and Meldolesi J: Shedding microvesicles: Artefacts no more. Trends Cell Biol. 19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Thery C, Amigorena S, Raposo G and Clayton A: Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. Chapter 3: Unit 3.22. 2006. View Article : Google Scholar | |
|
Musante L, Tataruch D, Gu D, Benito-Martin A, Calzaferri G, Aherne S and Holthofer H: A simplified method to recover urinary vesicles for clinical applications, and sample banking. Sci Rep. 4:75322014. View Article : Google Scholar : PubMed/NCBI | |
|
Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, et al: Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res (Amst). 4:214–222. 2010. View Article : Google Scholar | |
|
Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R: Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 6:69992015. View Article : Google Scholar : PubMed/NCBI | |
|
Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM and Simpson RJ: Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 56:293–304. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu L, Qu XH, Sun YL, Qian YM and Zhao XH: Novel method for extracting exosomes of hepatocellular carcinoma cells. World J Gastroenterol. 20:6651–6657. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Schageman J, Zeringer E, Li M, Barta T, Lea K, Gu J, Magdaleno S, Setterquist R and Vlassov AV: The complete exosome workflow solution: From isolation to characterization of RNA cargo. BioMed Res Int. 2013:2539572013. View Article : Google Scholar : PubMed/NCBI | |
|
Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A and Möller A: Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 4:270312015. View Article : Google Scholar : PubMed/NCBI | |
|
Eldh M, Lötvall J, Malmhäll C and Ekström K: Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol Immunol. 50:278–286. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, et al: Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 10:e01361332015. View Article : Google Scholar : PubMed/NCBI | |
|
Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, Salumets A and Peters M: Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 47:135–138. 2014. View Article : Google Scholar | |
|
Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O and Hendrix A: The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 3:32014. View Article : Google Scholar | |
|
El-Khoury V, Pierson S, Kaoma T, Bernardin F and Berchem G: Assessing cellular and circulating miRNA recovery: The impact of the RNA isolation method and the quantity of input material. Sci Rep. 6:195292016. View Article : Google Scholar : PubMed/NCBI | |
|
Li M, Rai AJ, DeCastro GJ, Zeringer E, Barta T, Magdaleno S, Setterquist R and Vlassov AV: An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods. 87:26–30. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Quackenbush JF, Cassidy PB, Pfeffer LM, Boucher KM, Hawkes JE, Pfeffer SR, Kopelovich L and Leachman SA: Isolation of circulating microRNAs from microvesicles found in human plasma. Methods Mol Biol. 1102:641–653. 2014. View Article : Google Scholar | |
|
Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al: Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 34:2690–2699. 2015. View Article : Google Scholar | |
|
Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, et al: Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 3:269132014. View Article : Google Scholar : PubMed/NCBI | |
|
Cvjetkovic A, Lötvall J and Lässer C: The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles. Mar 25–2014.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI | |
|
Gercel-Taylor C, Atay S, Tullis RH, Kesimer M and Taylor DD: Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal Biochem. 428:44–53. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Caradec J, Kharmate G, Hosseini-Beheshti E, Adomat H, Gleave M and Guns E: Reproducibility and efficiency of serum-derived exosome extraction methods. Clin Biochem. 47:1286–1292. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Webber J and Clayton A: How pure are your vesicles? J Extracell Vesicles. Jan 10–2013.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI | |
|
Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C and Le Pecq JB: Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 270:211–226. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Moret I, Sánchez-Izquierdo D, Iborra M, Tortosa L, Navarro-Puche A, Nos P, Cervera J and Beltrán B: Assessing an improved protocol for plasma microRNA extraction. PLoS One. 8:e827532013. View Article : Google Scholar : |