Sulforaphane reduces intracellular survival of Staphylococcus aureus in macrophages through inhibition of JNK and p38 MAPK‑induced inflammation
- Authors:
- Published online on: April 1, 2020 https://doi.org/10.3892/ijmm.2020.4563
- Pages: 1927-1941
-
Copyright: © Deramaudt et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
During bacterial infection, the innate immune system triggers a rapid and non-specific inflammatory response mechanism to prevent the spread of bacteria. As one of the first defense mechanisms used, the phagocytic macrophages serve an important role in clearing foreign particles, including bacteria and cell debris, by phagocytosis and by restoring cell homeo-stasis to prevent tissue damage (1,2). Following microbial or cell debris recognition, the acute inflammatory response is initiated and triggers the rapid release of proinflamma-tory mediators such as interleukin (IL)-1β, IL-6, and tumor necrosis factor-α (TNF-α), which will subsequently induce the adaptive immune system (3,4). In addition to their phagocytic activity, macrophages serve an essential role in the maintenance of tissue homeostasis. Furthermore, they exhibit an uncommon plasticity and are able to polarize into subsets of proinflammatory or anti-inflammatory populations depending on their surrounding environment and their own phagocytic activity (5). The mitogen-activated protein kinase (MAPK) signaling pathways, which include the most studied members ERK, p38 MAPK (p38) and JNK, are instrumental in all innate immune receptor signaling pathways. The MAPKs act by modulating the expression of inflammatory cytokines and chemokines (6,7).
Staphylococcus aureus, a gram-positive human bacterium, is a commensal opportunistic pathogen. It is a leading cause of skin and soft tissue infections, osteomyelitis, septic arthritis, pneumonia and endovascular infections (8). Unfortunately, treatments are complicated by the prevalence of multi-drug resistance strains, which result in high mortality and morbidity rates (9). Originally considered an extracellular pathogen, it has been demonstrated that S. aureus is also able to invade, survive and replicate within mammalian phagocytic and non-phagocytic cells using various survival strategies (10). A pathogenic characteristic of S. aureus is its ability to generate intense local and systemic inflammatory responses, evident in the release of inflammatory cytokines. Once released within the infected tissue environment, the cytokines act on the surrounding epithelial, stromal, and circulating cells, triggering secondary waves of cytokine release, consequently amplifying the host defense response. Uncontrolled levels of cytokines are detrimental to the host, resulting in massive tissue destruction.
Sulforaphane (SFN), a degradation product of gluco-sinolates, is an isothiocyanate derivative naturally present in cruciferous vegetables such as broccoli sprouts. SFN is a potent enhancer of phase II detoxification enzymes with chemopreventive, antitumor, anti-inflammatory and antimicrobial activities against a variety of bacterial and fungal species (11-13). While several in vitro studies have described SFN antimicrobial activity, SFN treatment in Helicobacter pylori infection was the most studied. This pathogenic bacteria is involved in gastric epithelium infection and is associated with gastric cancer, and at present, the molecular mechanism involved in the bactericidal effect of SFN against H. pylori remains elusive (11,14). Recently, treatment with SFN was demonstrated to inhibit HIV infection in THP-1-derived macrophages and primary macrophages, but not primary T cells (15). In this case, the inhibitory effect of SFN on microbial persistence is exerted through a nuclear factor erythroid 2-related factor 2 (Nrf2)-dependent mechanism. SFN is a well-known activator of the transcription factor Nrf2, a key regulator of genes coding for phase II detoxifying enzymes, including NAD(P)H quinone dehydrogenase 1 (NQO1) and heme oxygenase-1 (HO-1), cytoprotective, antioxidant and anti-inflammatory responses (16,17). Nrf2 is an essential factor in the attenuation of inflammation since Nrf2-deficient mice exhibit increased inflammation (18). In addition, Nrf2 has been suggested to negatively regulate the transcription of IL-1β and IL-6 proinflammatory cytokine genes in lipo-polysaccharide (LPS)-treated mouse bone marrow-derived macrophages (19).
Post-transcriptional regulation by microRNAs (miRNAs) has emerged as a major regulatory mechanism to control the expression level of genes involved in a number of fundamental cellular processes such as inflammation, proliferation, apoptosis and macrophage polarization upon pathogenic infection (20). miRNAs are molecules measuring 18-24 nucleotides in length and belong to the short non-coding RNA family. By binding to the 3′ untranslated region of their target mRNA, each mature miRNA is able to target up to hundreds of mRNAs and inhibit their expression (21). SFN and other glucosinolate derivatives have been demonstrated to modulate epigenetic alterations identified during carcinogenesis, including DNA methylation, histone modifications and miRNAs (22).
The present study investigated the underlying cellular mechanisms initiated by the pretreatment of human THP-1-derived macrophages, primary human peripheral blood mononuclear cells (PBMC)-derived macrophages, and primary mouse bone marrow-derived macrophages (BMDMs) with SFN and subsequent challenge with S. aureus. The effect of SFN pretreatment on the intracellular survival of S. aureus, macrophage apoptosis and macrophage inflammatory response was evaluated. Furthermore, Nrf2−/− BMDMs were used to assess whether the Nrf2 signaling was involved. Finally, the cellular signaling pathways, regulated by SFN and involved in the decrease in intracellular S. aureus survival, were examined using specific MAPK inhibitors. The results provided novel insights into the molecular mechanisms underlying the anti-inflammatory and anti-apoptotic activities mediated by SFN to disrupt S. aureus-favored environment in macrophages.
Materials and methods
Antibodies and reagents
SFN, phorbol 12-myristate 13-acetate (PMA), phalloidin-ATTO 594 (cat. no. 51927) were purchased from Sigma-Aldrich; Merck KGaA. The RPMI-1640 medium, heat-inactivated fetal bovine serum (FBS), FAM-FLICA caspases-3/7 assay kit (ImmunoChemistry Technologies, LLC) and human recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) were obtained from Eurobio Scientific. The Tryptic soy broth and Tryptic soy agar (Condalab) were obtained from Dominique Dutscher SAS. The Annexin V-fluorescein isothiocyanate apoptosis reagent, and the inhibitors SB203580 and SP600125 were obtained from Abcam. Rabbit anti-Nrf2 antibody (cat. no. 16396-1AP) was purchased from ProteinTech Group, Inc. Gentamycin sulfate, mouse anti-GAPDH (cat. no. CB1001), rabbit anti-phos-phorylated (p)-ERK (cat. no. 04-797), rabbit anti-ERK (cat. no. ABS44) and mouse anti-p-p38 (cat. no. MABS64) antibodies were obtained from Merck KGaA. Mouse anti-p-JNK (cat. no. 612540), mouse anti-JNK (cat. no. 610627), mouse anti-p38 (cat. no. 612168) antibodies were purchased from BD Biosciences. The iTaq SYBR-Green Supermix and DC protein assay kit were purchased from Bio-Rad Laboratories, Inc. Goat anti-rabbit IRDye 680RD (cat. no. 926-68071) and goat anti-mouse IRDye 800CW (cat. no. 926-32210) antibodies were obtained from LI-COR Biosciences. Specific miRCURY LNA miRNA PCR primer sets (hsa-miR-142-5p, has-miR-146a-5p and UniSp6), and the miRCURY LNA RT and miRCURY LNA SYBR PCR kits were purchased from Qiagen. Dulbecco's modified Eagle medium (DMEM), Maxima First Strand cDNA synthesis kit, CellRox Green flow cytometry assay kit and Ficoll-Paque™ (GE Healthcare) were obtained from Thermo Fisher Scientific, Inc.
Animals
Breeding of the C57BL/6J Nrf2 knockout (Nrf2−/−) mouse strain, provided by Dr Yamamoto (Tohuki University) and purchased from Riken BRC (17), was performed at the University of Versailles-Saint-Quentin-en-Yvelines (license no. APAFIS8785-201610191723731v3). The protocols were approved by the Animal Ethics Committee CEEA47 PELVIPHARM of the University of Versailles-Saint-Quentin-en-Yvelines and the Ministry of Higher Education, Research and Innovation. Nrf2 heterozygote (Nrf2+/−) littermates were used as the control group.
Cell culture, cell differentiation and treatments
The human THP-1 monocytic cell line [American Type Culture Collection (ATCC®) TIB-202™] was maintained in RPMI-1640 supplemented with 10% heat-inactivated FBS, 1 mM sodium pyruvate, 10 mM HEPES and 0.05 mM β-mercaptoethanol in a humidified atmosphere at 37°C and 5% CO2. Terminal differentiation of THP-1 to macrophages was obtained by rinsing the cells twice with sterile PBS prior to incubation at 37°C with 50 nM PMA for 48 h.
To obtain the PBMC-derived macrophages, human primary monocytes were isolated from heparinized peripheral blood purchased from the Etablissement Français du Sang (EFS) using a Ficoll-Paque™ gradient. Blood samples were diluted with an equal volume of PBS and centrifuged at room temperature for 30 min at 400 x g. The EFS authorized the Inserm U1179 research unit (agreement number 15/EFS/012) to use anonymized blood samples for non-therapeutic purposes. PBMCs were cultured in the abovementioned culture medium without β-mercaptoethanol with the addition of 20 ng/ml GM-CSF. A total of 7 days after isolation, non-adherent cells were removed and adherent PBMC-derived macrophages were ready for treatment.
For the BMDMs isolated from Nrf2−/− and Nrf2+/− littermate mice, bone marrow was extracted from mouse femurs and tibias, washed with PBS and cultured in DMEM supplemented with 20% filtered L929 fibroblast cell line-conditioned media and 10% heat-inactivated FBS in non-treated culture dishes. Conditioned medium was harvested from L929 fibroblasts, grown in DMEM supplemented with 10% heat-inactivated FBS, and 3 days following confluency it was filtered through a 0.22-µm filter and stored at -20°C. A total of 7 days after harvest, all non-adherent cells were removed, and the remaining macrophages were detached by resuspending cells in cold PBS, and by scraping cells after 10 min incubation on ice. BMDMs were stained 1 min at room temperature by diluting the cell suspension with an equal volume of 0.4% trypan blue, counted using the Countess automated cell counter (Thermo Fisher Scientific, Inc.), and seeded in 24-well culture plates at a density of 5×105/well for use the following day.
When required, macrophages were incubated at 37°C with p38 or JNK inhibitors (25 µM SB203580 and 25 µM SP600125, respectively) 1 h prior to addition of 10 µM SFN (23). A total of 3 h after SFN treatment, S. aureus was added at a multiplicity of infection (MOI) of 10, and the cells were incubated at 37°C for an additional 3 h prior to cell lysis and RNA isolation. For 24 h post-infection assays, 20 µg/ml gentamycin was added to cell medium 1 h after S. aureus infection, to eliminate extracellular bacteria.
Total RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated 3 h after S. aureus infection of macrophages with TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and chloroform extraction technique following the manufacturer's protocol. Total RNA was quantified using the GE NanoVue spectrophotometer (GE Healthcare). To determine mRNA expression levels of target genes HO-1, NQO1, IL-1β, IL-6, TNF-α, C-C motif chemokine receptor 7 (CCR7), IL-23, iNOS, and 8S ribosomal RNA, 1 µg total RNA was reverse transcribed to cDNA using Maxima First strand cDNA synthesis kit following the manufacturer's protocol. cDNAs were then analyzed by RT-qPCR using iTaq SYBR-Green qPCR mix. The thermo-cycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles of a 5 sec denaturation step at 95°C and a 20 sec annealing/extension step at 60°C. The specific oligonucleotides for human and mouse HO-1, NQO1, IL-1β, IL-6, TNF-α, 18S ribosomal RNA, CCR7, IL-23 and iNOS, listed in Table I, were synthetized by Eurogentec. Gene expression levels were normalized to that of the reference gene 18S ribosomal RNA. To analyze miRNA expression levels of target genes miR-142-5p and miR-146a-5p, miRNA was reverse transcribed using the miRCURY LNA RT kit. cDNAs were then analyzed by qPCR using miRCURY LNA SYBR PCR kit. miRNA expression levels of target genes were normalized to the internal control U6 snRNA level. RT-qPCR was performed using the CFX96 and CFX384 Touch thermocyclers (Bio-Rad Laboratories, Inc.), with each sample performed in triplicate. Data were analyzed on BioRad CFX manager 3.1 using the 2−ΔΔCq method (24). Each RT-qPCR assay was performed independently and in triplicate.
Bacterial strain, growth culture, and fluorescence labeling of S. aureus
The ATCC 25923TM strain was grown aerobically in Tryptic soy broth to an optical density of 1 at 37°C under agitation. Bacterial glycerol stocks were maintained at -20°C. When required, frozen stocks were thawed and bacteria were diluted to the appropriate inoculum in sterile PBS. For the intracellular bacterial survival assay, bacteria were grown on Tryptic soy broth solidified with 1.5% agar. For the S. aureus internalization assay, fresh fluorescent S. aureus were generated by incubating S. aureus with SYTO9 for 15 min (Thermo Fisher Scientific, Inc.) at room temperature in the dark under gentle shaking, then washing 3 times in sterile PBS, and resus-pending in PBS prior to cell infection.
S. aureus internalization assay and immunofluorescence labeling
THP-1 cells were seeded in 6-well plates at 1×106 cells/well and differentiated with 50 nM PMA for 48 h. THP-1-derived macrophages were infected with SYTO9-labeled S. aureus at an MOI of 10. After 1 h infection, cells were washed twice with cold PBS to prevent additional bacterial internalization and to remove extracellular bacteria. Cells were then trypsinized at 37°C for 10 min, fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature, and finally suspended in 500 µl 0.02% EDTA. The quantification of cells infected with SYTO9-labeled S. aureus were performed using BD LSRFortessa flow cytometer and analyzed using FACSDiva software 7.0 (BD Biosciences). A forward and side scatter gate was set to exclude dead and aggregated cells. A total of 100,000 events per condition was collected.
For the immunofluorescence labeling, THP-1-derived macrophages seeded at 5×105 cells/coverslip were infected with SYTO9-stained S. aureus for 1 h then cells were rinsed with PBS and fixed for 10 min at room temperature with 4% PFA. Cells were visualized by labeling with phalloidin-ATTO 594. Images were captured using a laser scanning confocal fluorescence microscope (Leica SP8; Leica Microsystems, Inc.) and analyzed using ImageJ v.1.47 software (National Institutes of Health).
Intracellular survival assay of S. aureus
For the intracellular bacterial survival assay, macrophages seeded in 24-well plates at 2.5×105 cells/well were infected with S. aureus at an MOI of 10. After 1 h infection, cells were washed with PBS and extracellular bacteria were eliminated by the addition of 20 µg/ml gentamycin in cell culture medium. After 24 h incubation at 37°C, cells were rinsed with PBS and lysed in 1 ml ice-cold sterile water for 20 min at 4°C. Intracellular bacteria were diluted in 5-fold serial dilutions and then plated on Tryptic soy agar plates. Colony forming units (CFU) counts were determined 24 h after incubation at 37°C.
Quantification of reactive oxygen species (ROS) production
The CellRox green reagent assay kit (Thermo Fisher Scientific, Inc.) was used to determine ROS levels. Briefly, THP-1-derived macrophages were seeded at a density of 1×105 cells/well on coverslip-covered wells in 24-well plates. Following treatment, macrophages were stained with 5 µM CellRox green reagent for 30 min at 37°C. The cells were then washed with PBS, fixed in 4% PFA for 15 min at room temperature and the nuclei were stained with 10 µg/ml 4',6-diamidino-2-phe-nylindole dihydrochloride for 10 min at room temperature. Analysis of the images captured with the Leica SP8 confocal microscope (Leica Microsystems, Inc.) were performed using ImageJ v.1.47 software (National Institutes of Health).
Cell viability assay
THP-1-derived macrophages were seeded at 5×104 cells/well in a 96-well plate. Cytotoxicity to SFN or S. aureus infection was assessed by an MTT cell viability assay kit (Biotium, Inc.) to measure cellular metabolic activity following the manufacturer's protocol. Absorbance changes were measured at 550 nm, and the background absorbance at 600 nm, using the FLUOstar Omega microplate reader (BMG Labtech).
Caspases-3/7 activity assays
When indicated, BMDMs seeded in 6-well plates at 1×106 cells/well were pretreated for 3 h with 10 µM SFN or DMSO prior to infection with S. aureus (MOI of 10). Gentamycin 20 µg/ml was added to the culture medium 1 h after infection to eliminate extracellular S. aureus. Staurosporine (1 µM) was used as an apoptosis positive control. Following cell treatment, caspases-3/7 activities were determined using the Green FAM-FLICA Caspases-3/7 assay kit according to the manufacturer's protocol. Briefly, the cells were trypsinized and incubated at 37°C in the dark for 1 h with the FLICA probe, made of the irreversible caspase inhibitor DEVD-fluoromethyl ketone fused to a carboxyfluorescein (FAM-DEVD-FMK), which binds specifically and covalently to activated caspase-3/7 enzymes. Following 2 washes, cells were fixed using 4% PFA for 10 min at room temperature and resuspended in 0.02% EDTA and the number of FLICA-positive cells was counted using the BD LSRFortessa™ flow cytometer and analyzed using FACSDiva 7.0 software (BD Biosciences).
Protein extraction and western blot analysis
For total protein extraction, THP-1-derived macrophages were rinsed with cold PBS and then lysed with cold RIPA buffer [150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris-HCl (pH 7.5)], supplemented with protease and phosphatase inhibitor cocktail mixtures. Protein concentrations were determined using the DC protein assay kit. Total proteins (20 µg/lane) were resolved by 4-20% SDS-PAGE and transferred to Immobilon-FL polyvinylidene difluoride membranes (Merck KGaA). Western blot analysis was then performed at room temperature for ~3 h using the iBind Flex Western system (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Briefly, primary antibodies against Nrf2 (1:1,000), GAPDH (1:8,000), totaland p-ERK (1:2,000 and 1:1,000, respectively), total and p-p38 (1:2,000 and 1:1,000, respectively), total and p-JNK (both 1:1,000), and IRDye680RD- or IRDye800RD-conjugated secondary antibodies (1:4,000 and 1:3,000, respectively) were diluted in iBind Flex FD Solution. Fluorescent blot imaging was performed using Odyssey CLx Imaging system (LI-COR Biosciences). Densitometric analysis was performed using Image Studio Lite software v4.0 (LI-COR Biosciences).
Statistical analysis
The data are presented as the mean ± standard error of the mean (SEM) from at least 3 independent experiments. Imaging flow cytometry data are presented as the mean ± SEM of at least 3 independent experiments analyzed from 100,000 events each. Statistical comparisons were performed using two-tailed unpaired t-tests or ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
SFN improves intracellular S. aureus clearance in macro- phages
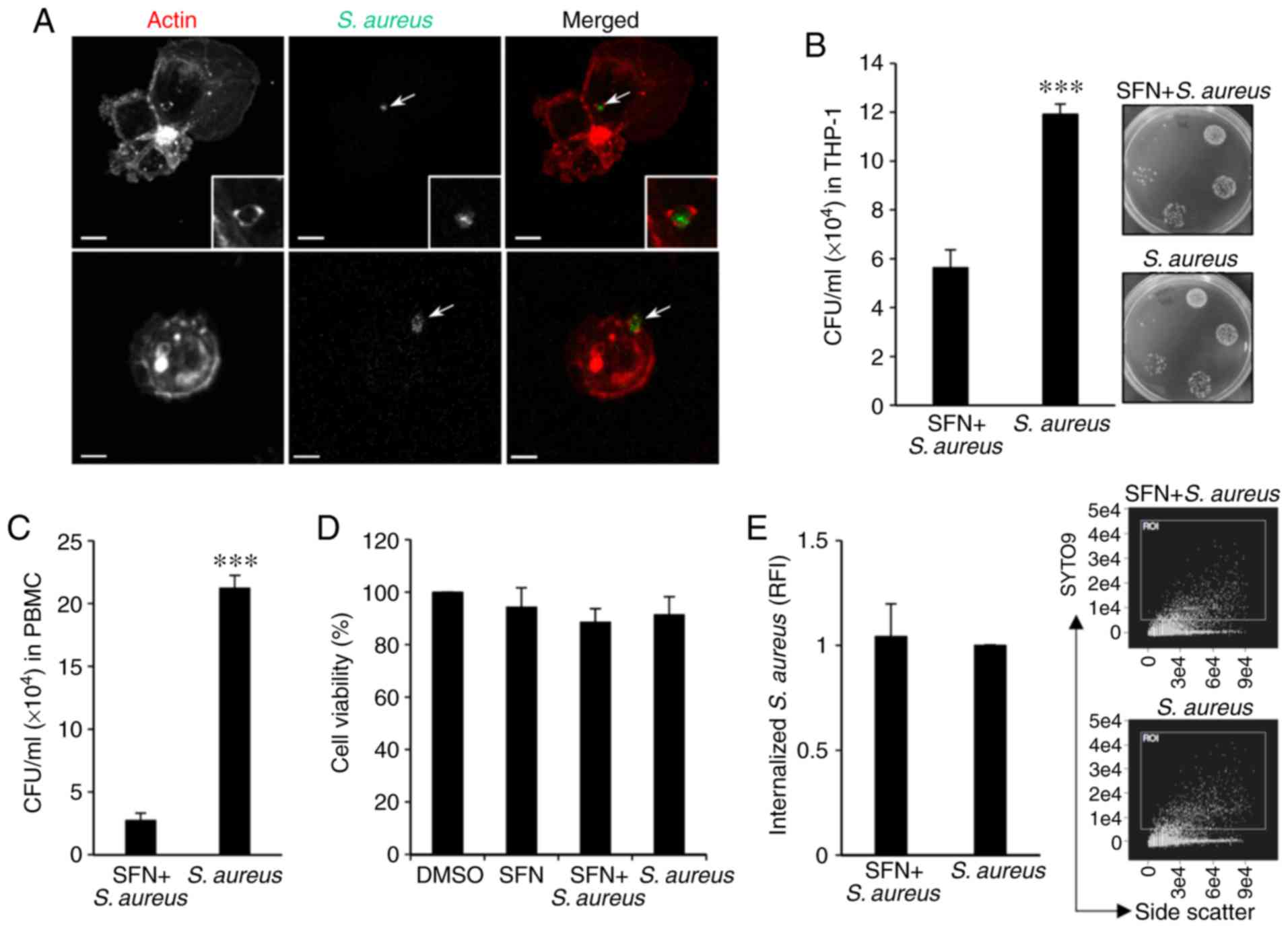
In order to gain a better understanding of the effect of SFN on the intracellular fate of S. aureus in macrophages, THP-1-derived macrophages were utilized. To visualize the internalization of S. aureus, THP-1 monocytes were first differentiated into macrophages with PMA and infected with SYTO9-stained S. aureus for 1 h. Following PFA fixation, the macrophages, stained with fluorescent phalloidin and intracellular S. aureus, were visualized by confocal microscopy. Analysis of the images of infected macrophages demonstrated the internalization of SYTO9-labeled S. aureus in THP-1-derived macrophages, including S. aureus enclosed in actin-dependent structures reminding of early phagocy-tosis (Fig. 1A).
To evaluate the intracellular survival of S. aureus, THP-1-derived macrophages were pretreated for 3 h with 10 µM SFN or the vehicle DMSO prior to S. aureus infection. A total of 1 h after infection, gentamycin was added to the culture medium to eliminate extracellular bacteria and infected macrophages were incubated for an additional 24 h post-infection. CFU enumeration indicated that intracellular bacteria counts were significantly decreased in SFN pretreated THP-1-derived macrophages compared with the DMSO pretreated cells (Fig. 1B). The inhibitory effect of SFN observed in the THP-1-derived macrophages was confirmed by infecting macrophages derived from human peripheral blood mononuclear cells (PBMCs). PBMCs issued from blood donors and isolated using a Ficoll-Paque™ gradient were first differentiated to macrophages for 7 days with GM-CSF prior to pretreatment with SFN or DMSO and infection with S. aureus. SFN pretreated PBMC-derived macrophages exhibited a significant decrease in intracellular bacterial survival 24 h after infection compared to S. aureus counts issued from DMSO treated macrophages (Fig. 1C).
Furthermore, to determine whether cytotoxicity contributed to the apparent decrease in bacterial persistence, cell viability was assessed in THP-1-derived macrophages incubated with SFN or infected for 1 h with S. aureus prior to the addition of gentamycin and 24 h incubation. The MTT assay revealed that neither SFN nor S. aureus affected THP-1-derived macrophage viability as compared with that observed in the DMSO-treated cells (Fig. 1D). Additionally, quantification by flow cytometry of internalized SYTO9-stained S. aureus in THP-1-derived macrophages 1 h after infection indicated that SFN pretreatment of macrophages had no effect on S. aureus internalization when compared with the DMSO-treated macrophages infected with S. aureus (Fig. 1E). The modulation of the oxidative stress by S. aureus in THP-1-derived macrophages was also assessed. ROS levels were measured in macrophages 24 h after bacterial challenge in presence of the CellRox fluorogenic probe. Confocal images and fluorescent intensities were analyzed by ImageJ software, and the results indicated that there was no significant modification to ROS production in THP-1-derived macrophages pretreated with SFN and/or infected with S. aureus compared with the DMSO-treated macrophages (Fig. S1), indicating that the SFN-mediated inhibition of intracellular S. aureus survival in THP-1-derived macrophages was not due to an effect of SFN or S. aureus on cell viability or ROS production.
SFN affects intracellular S. aureus survival and cell apoptosis in an Nrf2-independent manner
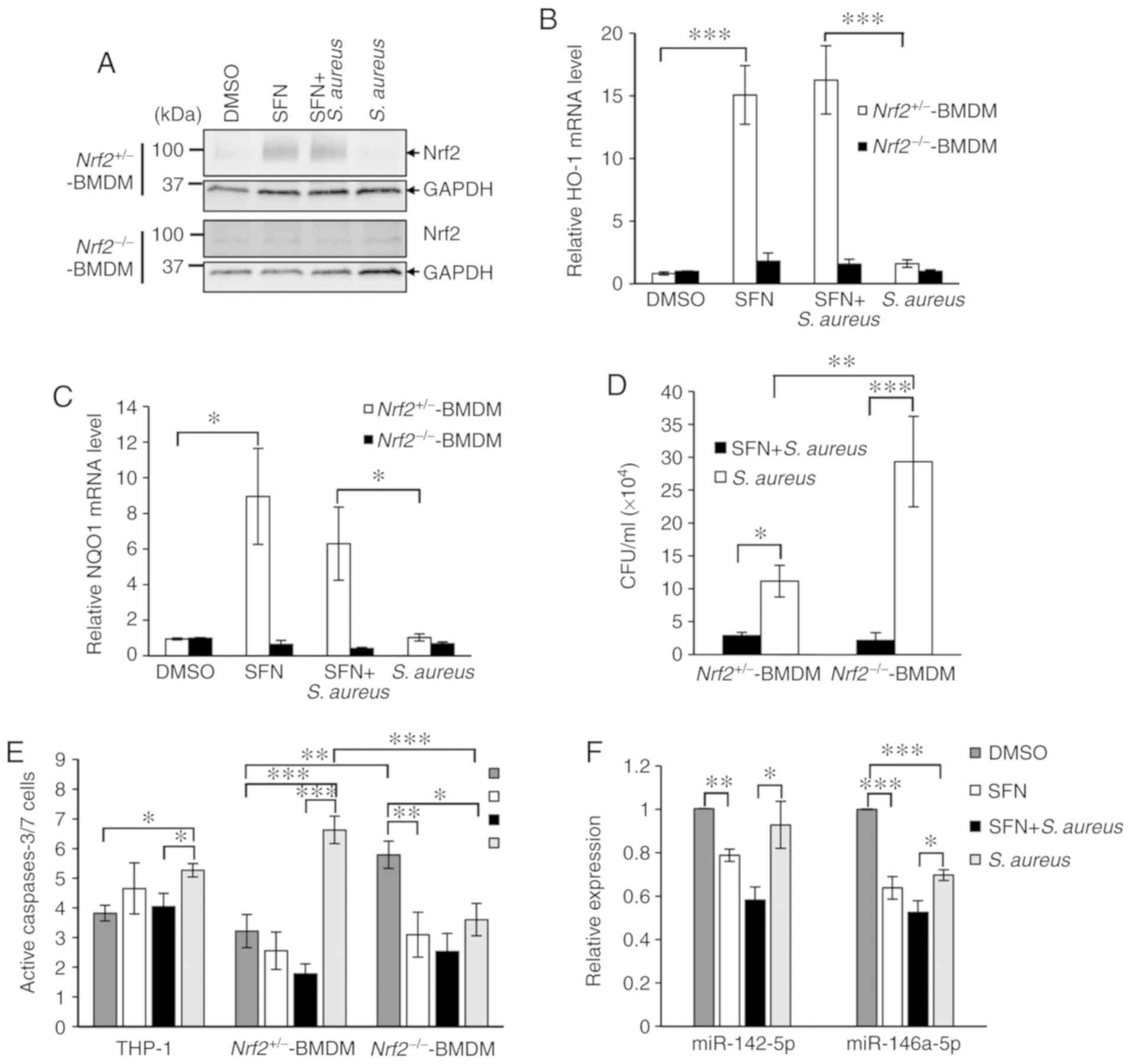
As SFN is a well-known activator of transcription factor Nrf2, a key regulator of inducible intracellular defenses in the innate immune system, the present study aimed to determine whether SFN activated Nrf2 signaling to impair intracellular survival of S. aureus in macrophages. Primary bone marrow stem cells were isolated from Nrf2−/− mice and their control Nrf2+/− littermates, and differentiated into BMDMs using L929-conditioned media. Nrf2+/− and Nrf2−/− BMDMs pretreated with DMSO or SFN were infected with S. aureus for 1 h prior to the addition of gentamycin to the culture medium. Total proteins were extracted 24 h after infection and examined by western blot analysis. SFN significantly increased the Nrf2 protein levels in both non-infected and infected Nrf2+/− BMDMs compared with the DMSO-treated macrophages, whereas no detectable Nrf2 was observed in the non-infected and infected macrophages pretreated with DMSO (Fig. 2A). Knockout of Nrf2 in the Nrf2−/− BMDMs was validated by western blot analysis and qPCR analysis of 2 target genes of Nrf2 coding for HO-1 and NQO1. qPCR analysis demonstrated the lack of transcription upregulation of HO-1 and NQO1 in DMSO-treated or S. aureus-infected Nrf2+/− BMDM, and in Nrf2−/− BMDMs pretreated with SFN (Fig. 2A-C).
The effect of Nrf2 and Nrf2-knockout on the intracellular survival of S. aureus in Nrf2−/− and Nrf2+/− BMDMs pretreated with SFN or DMSO was next investigated. Similar to THP-1-derived macrophages and primary PBMC-derived macrophages (Fig. 1B and C), Nrf2+/− BMDMs exhibited a significant decrease in intracellular bacterial survival when pretreated with SFN compared with Nrf2+/− BMDMs pretreated with DMSO (Fig. 2D). Noticeably, S. aureus survival levels were significantly decreased following SFN pretreatment in Nrf2−/− BMDMs, suggesting that the effect of SFN on bacterial survival was independent of Nrf2. Furthermore, bacterial survival was increased in the S. aureus-infected Nrf2−/− BMDMs compared with the S. aureus-infected Nrf2+/− BMDM, which suggested a role of Nrf2 signaling in moderating intracellular S. aureus survival in macrophages, independently of SFN.
Following S. aureus infection, immune cells can undergo caspase-dependent cell death to contribute to the control of infection. The resulting apoptotic bodies, comprised of endo-cytosed pathogens and antigen presenting cell debris, facilitate T-cell response and clearance by neighboring immune cells (25). To verify the effect of SFN on S. aureus mediated caspase-3/7 activity, the FAM-FLICA caspase-3/7 assay kit was used on THP-1-derived macrophages or BMDMs pretreated with SFN or DMSO and infected with S. aureus. Gentamycin was added to the medium 1 h after infection and cells were incubated for 24 h post-infection. Apoptosis analysis by flow cytometry indicated that S. aureus significantly activated caspases-3/7 in THP-1-derived macrophages and Nrf2+/− BMDMs, while the lack of Nrf2 in S. aureus-challenged Nrf2−/− BMDMs resulted in a significant decrease in S. aureus-mediated caspase-3/7-dependent cell apoptosis as compared with the DMSO-treated Nrf2−/− BMDMs and S. aureus-infected Nrf2+/− BMDMs (Fig. 2E). In addition, an increase in caspases-3/7 activity was detected in the DMSO-treated Nrf2−/− BMDMs as compared with that measured in the DMSO-treated Nrf2+/− BMDM, which was inhibited by SFN pretreatment. SFN pretreatment of the THP-1-derived macrophages and Nrf2+/− BMDMs decreased the level of S. aureus-triggered caspases-3/7-dependent cell apoptosis. Notably, this negative effect of SFN was maintained in the S. aureus-infected Nrf2−/− BMDM. The expression levels of miR-142-5p and miR-146a-5p have been associated with cell apoptosis (26,27). Therefore, the effects of SFN on the levels of these miRNAs were examined. RT-qPCR analyses demonstrated that SFN pretreatment of the THP-1-derived macrophages significantly attenuated the S. aureus-mediated increased expression levels of miR-142-5p and miR-146a-5p (Fig. 2F). Altogether, these results suggested a negative correlation between decreased S. aureus-mediated cell apoptosis in Nrf2−/− BMDMs and increased intracellular bacterial survival. Although Nrf2 clearly participates in the control of S. aureus intracellular persistence, the inhibitory effects of SFN on macrophage bactericidal activity and caspase-dependent cell apoptosis appears to be Nrf2-independent.
SFN inhibits S. aureus-induced transcriptional expressions of proinflammatory genes
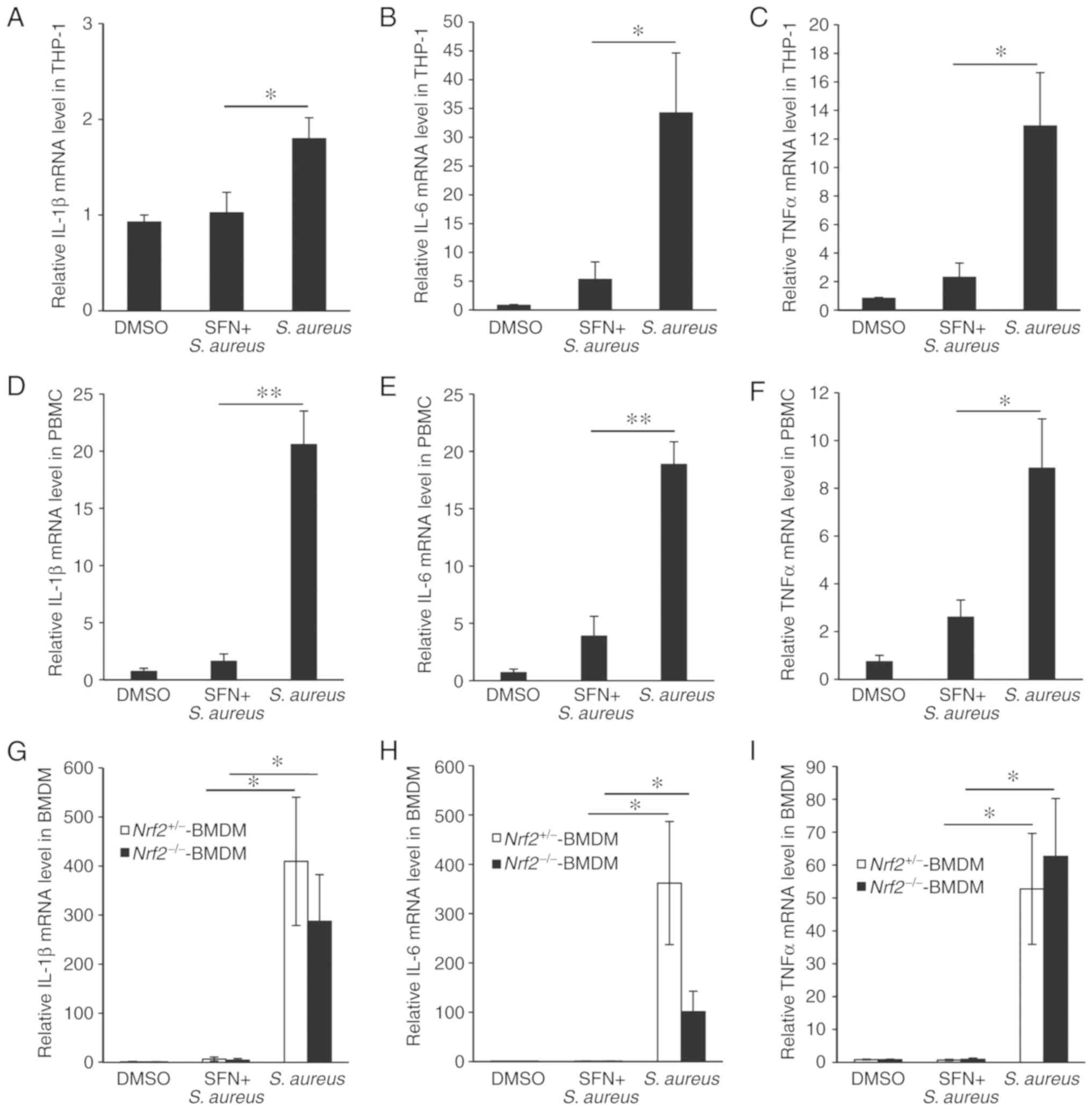
The inflammatory response triggered by S. aureus infection in macrophages and the effect of SFN on S. aureus-induced inflammation were then examined. THP-1-derived macrophages were pretreated with SFN or DMSO and infected with S. aureus. Total RNAs were extracted 3 h after infection and the transcription levels of the genes coding for proinflammatory cytokines IL-1β, IL-6 and TNF-α were measured by RT-qPCR. The levels of IL-1β, IL-6 and TNF-α were significantly increased in the S. aureus-infected THP-1-derived macrophages compared with the non-infected macrophages (Fig. 3A-C). Pretreatment with SFN prior to bacterial challenge prevented S. aureus-mediated increase in IL-1β, IL-6 and TNF-α mRNA expression levels. Similar inhibitory effect of SFN was observed in primary PBMC-derived macrophages infected with S. aureus (Fig. 3D-F).
As Nrf2 has been recently demonstrated to directly affect the transcriptional induction of proinflammatory cytokine genes inflammation in macrophages, the participation of Nrf2 in the regulation of SFN-mediated increase was investigated (19). S. aureus significantly increased the transcriptional expression levels of IL-1β, IL-6 and TNF-α genes in Nrf2+/− and Nrf2−/− BMDM, and pretreatment of BMDMs with SFN prevented S. aureus-mediated transcription of the proinflammatory cytokine genes in both macrophages expressing Nrf2 and lacking Nrf2 (Fig. 3G-I), suggesting that SFN inhibited S. aureus-induced inflammation independently of Nrf2.
SFN prevents the S. aureus-induced expression of M1 marker genes in infected macrophages
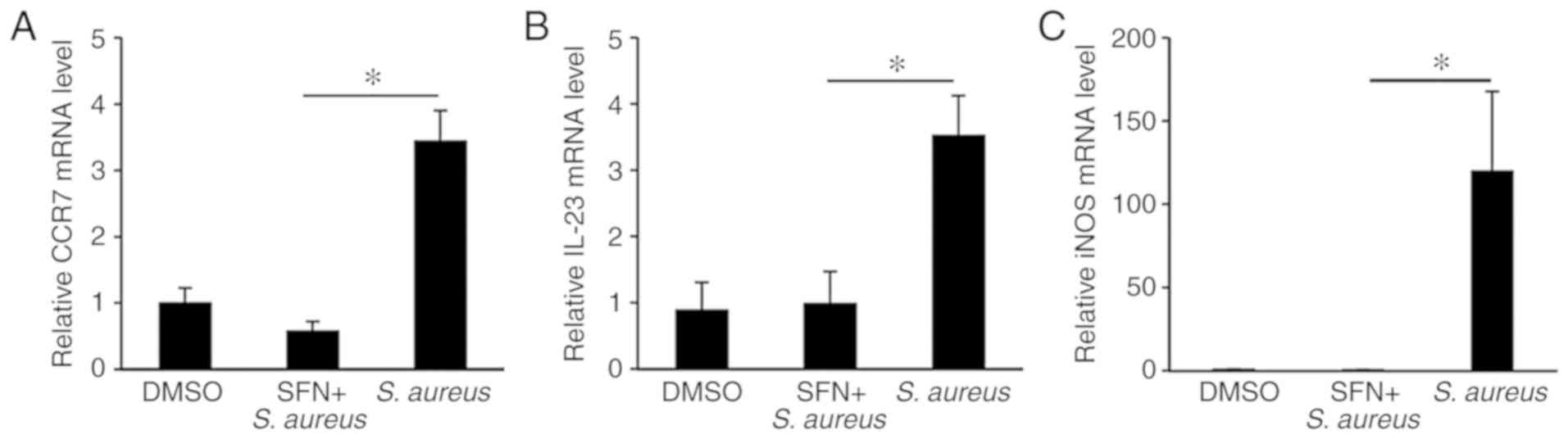
Upon bacterial infection, activated macrophages produce M1-like proinflammatory marker genes including IL-1β, IL-6 and TNF-α (28). The upregulation of M1 marker genes CCR7 and IL-23 genes in the THP-1-derived macrophages and the iNOS gene in the Nrf2+/− BMDMs following S. aureus infection was then confirmed by qPCR (Fig. 4A-C). SFN pretreatment of macrophages attenuated the levels of S. aureus-induced M1 marker genes to those observed in non-infected macrophages. These results suggest that pretreatment with SFN thwarts the classical activation of macrophages by S. aureus.
SFN modulates the MAPK signaling pathway
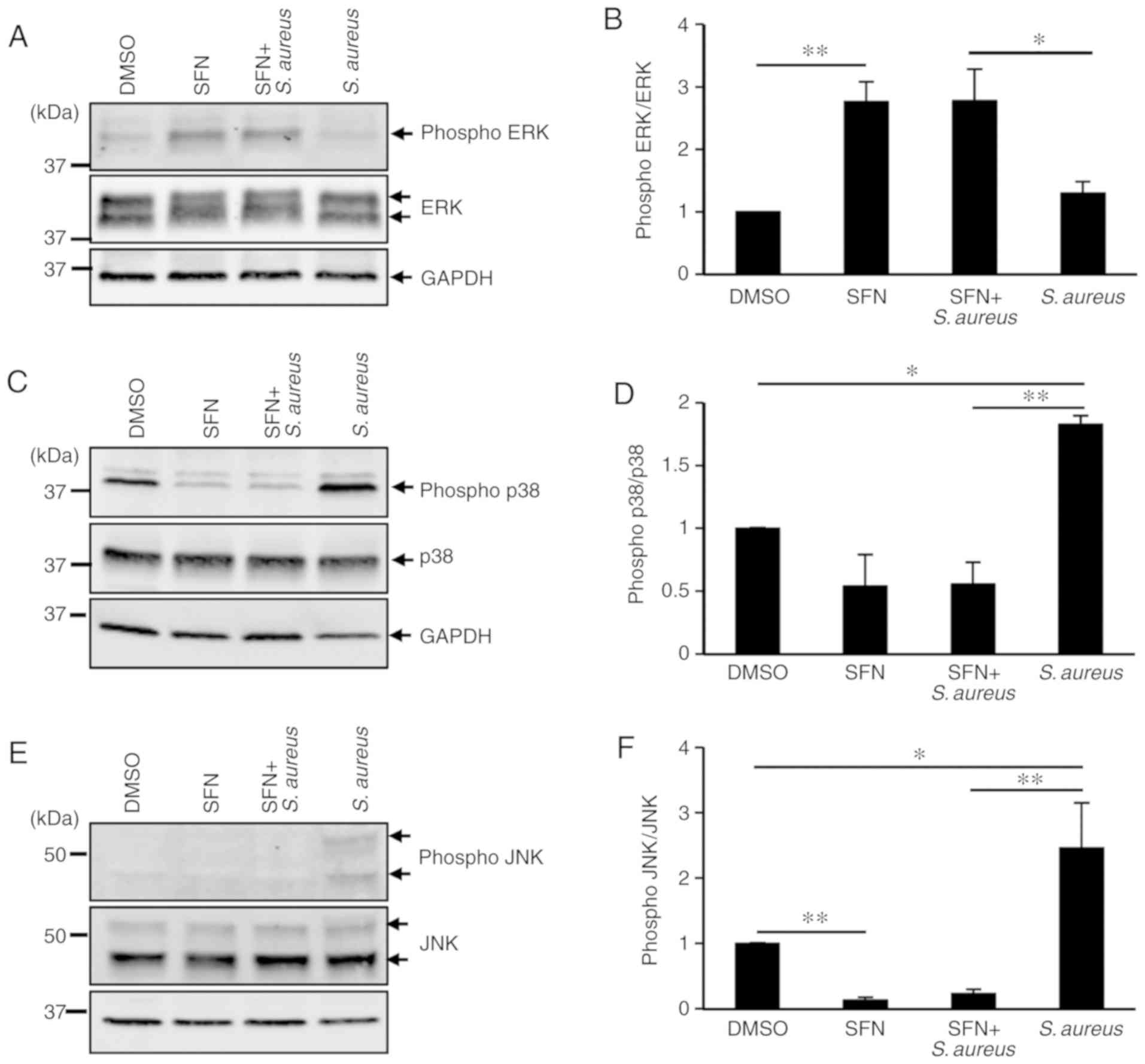
To investigate the molecular mechanisms employed by SFN to markedly decrease S. aureus-induced inflammation, the present study focused on the MAPK signaling pathways, as MAPK cascades are significantly involved in the macrophage inflammatory responses. ERK, p38 and JNK are the 3 most studied members of the MAPK family. To determine whether S. aureus activated ERK, p38 and JNK, THP-1-derived macrophages were pretreated with DMSO or SFN followed by S. aureus infection. Protein extracts were examined by western blot analysis, and the phosphorylation states of ERK, p38 and JNK were determined. Densitometry analysis of total ERK and phosphorylated ERK protein signals demonstrated a significant increase in phosphorylated ERK in SFN-treated macrophages compared with the DMSO-treated macrophages (Fig. 5A and B). No modification in the activation of the ERK pathway was observed in the S. aureus-challenged macrophages, while phosphorylation of p38 and JNK was significantly augmented (Fig. 5C-F). SFN pretreatment of THP-1-derived macrophages notably prevented activation of p38 and JNK in S. aureus infected and non-infected macrophages. These results suggested a strong inhibitory effect of SFN on the p38 and JNK signaling pathways.
MAPK-dependent inhibition of IL-1β, IL-6 and TNF-α mRNA expression levels by SFN
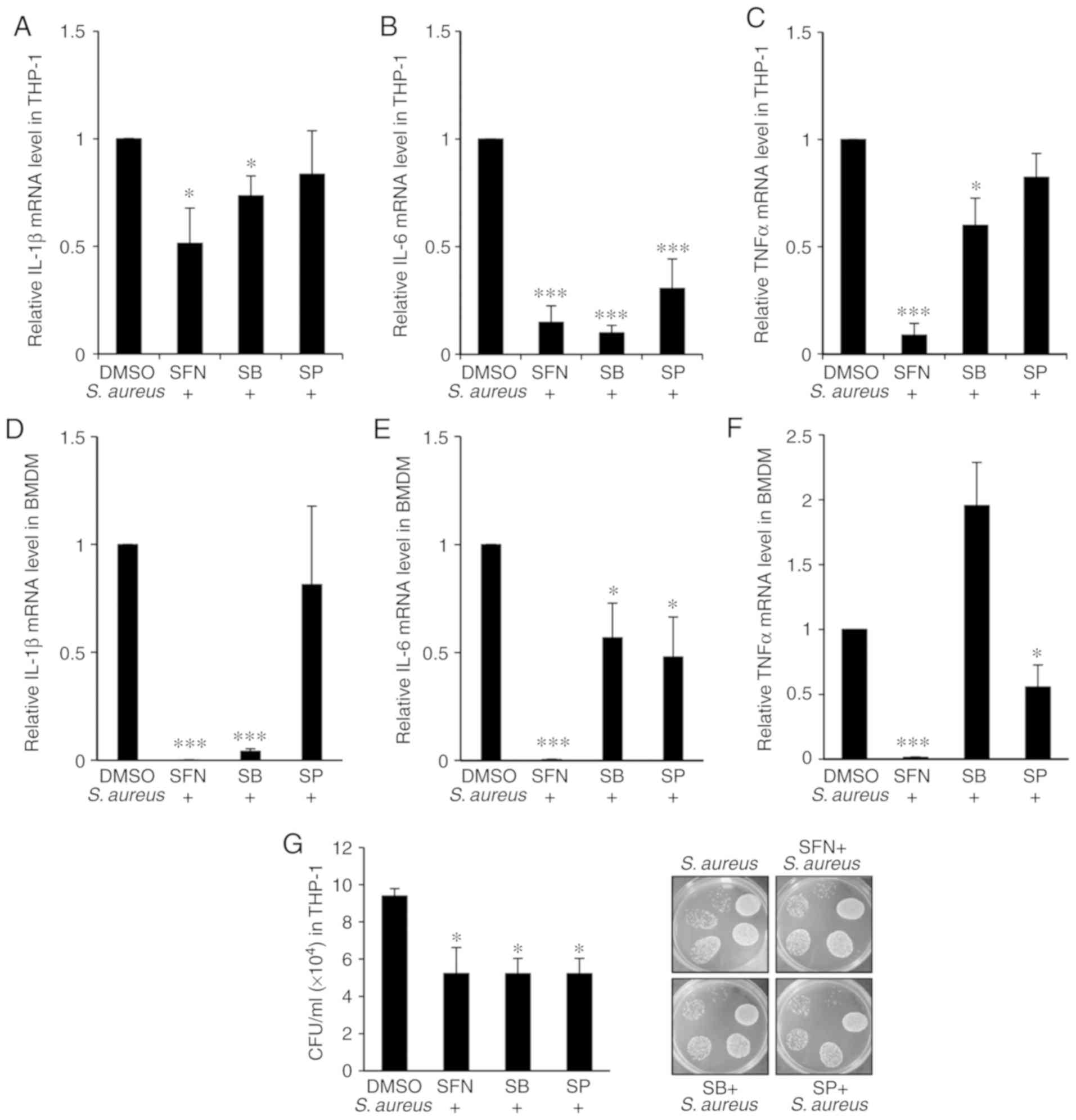
To further investigate whether the anti-inflammatory effect of SFN on the proin-flammatory cytokine genes IL-1β, IL-6 and TNF-α resulted from the inhibition of p38 or JNK phosphorylation by SFN, specific p38 and JNK inhibitors were used. THP-1-derived macrophages were pretreated with the p38 inhibitor SB203580 or the JNK inhibitor SP600125 prior to SFN treatment and/or S. aureus infection. qPCR analysis indicated that SB203580 effectively repressed the S. aureus-mediated increase in IL-1β, IL-6 and TNF-α gene expression levels when compared with the DMSO-pretreated macrophages infected with S. aureus (Fig. 6A-C). In addition, the decrease in IL-1β and IL-6 gene expression levels by SB203580 was comparable with that observed in the SFN-pretreated macrophages challenged with S. aureus. SP600125 significantly decreased the expression levels of IL-6 in the THP-1-derived macrophages infected with S. aureus compared with the macrophages pretreated with DMSO and infected with S. aureus (Fig. 7B). Data obtained from the Nrf2+/− BMDMs validated the inhibitory effect of SB203580 on IL-1β and IL-6 gene transcription (Fig. 6D and E), while SP600125 specifically inhibited the transcription of IL-6 and TNF-α genes (Fig. 6E and F).
As SFN suppressed p38 and JNK phosphorylation it was of interest to examine whether pretreatment of macrophages with SB203580 or SP600125 resulted in an attenuation of S. aureus load in macrophages. The THP-1-derived macrophages were pretreated with SFN, SB203580 or SP600125 and challenged with S. aureus for 1 h prior to the addition of gentamycin. CFU counts were determined after 24 h, and a significant decrease in intracellular bacterial survival in macrophages treated with SB203580 or SP600125 was observed, at a level comparable with that obtained in SFN pretreated macrophages (Fig. 6G). Overall, the results suggested that SFN activation of macrophage bactericidal activity is based on the inhibition of the macrophage proinflammatory responses by inhibiting p38 and JNK MAPK signaling.
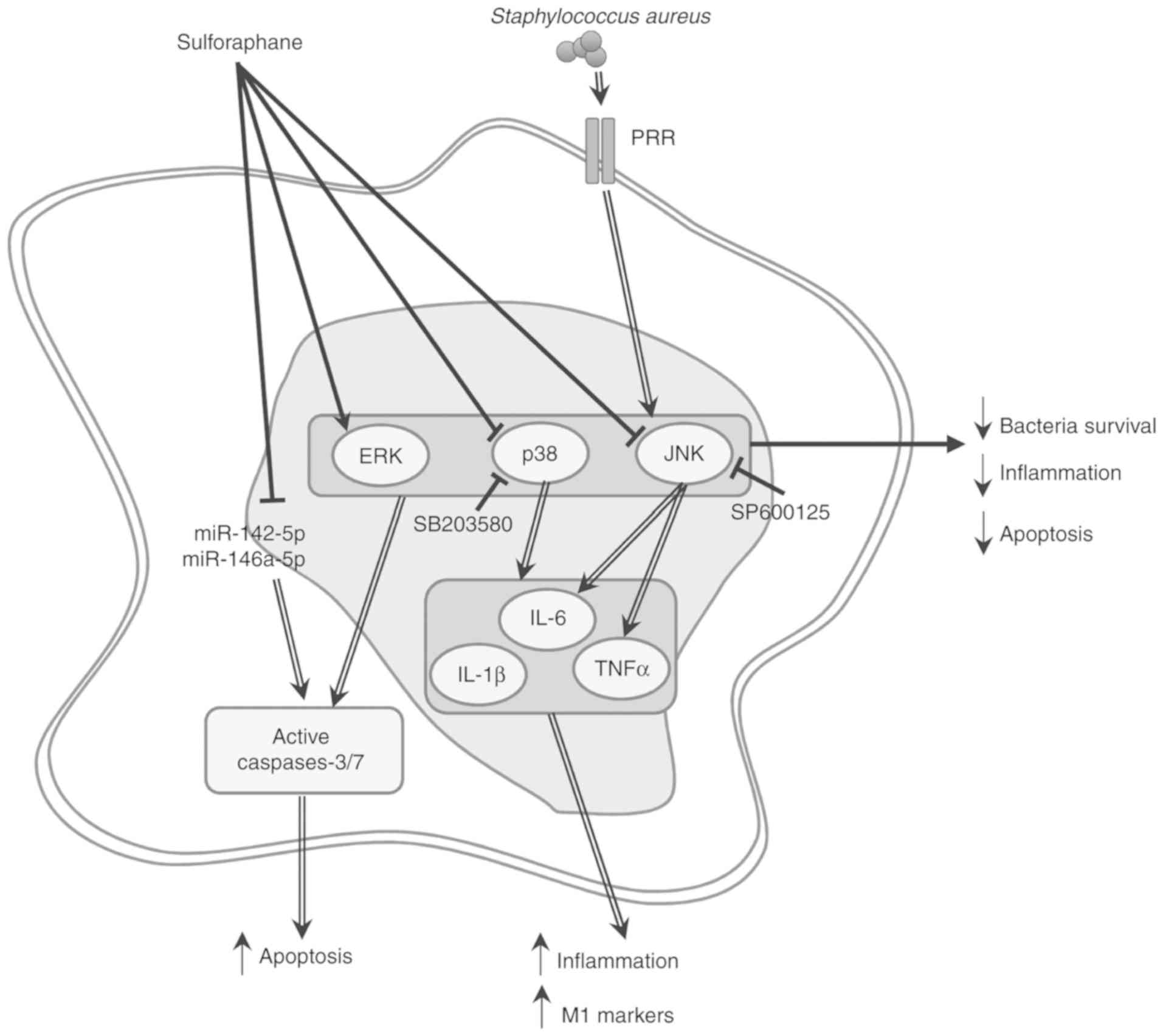
Fig. 7 summarizes the hypothesis regarding the role of SFN in inhibiting S. aureus recognition, but not its internalization, thereby preventing p38 and JNK MAPK signaling, and inhibiting MAPK-dependent upregulation of IL-1β, IL-6 and TNF-α proinflammatory cytokine genes, as well as potentially averting macrophage M1 polarization.
Discussion
SFN, a naturally-occurring isothiocyanate present in abundance in cruciferous vegetables such as broccoli sprouts, has been demonstrated to possess pleiotropic protective effects against oxidative stress, various types of cancer, inflammatory diseases and microbial infections (11-13). In the present study, the molecular mechanism activated by SFN to improve bacterial clearance in macrophages challenged with S. aureus was identified. It was demonstrated that SFN decreases intracellular bacterial survival by preventing the activation of p38 and JNK signaling pathways activated by S. aureus, 2 major enzymes implicated in the production of inflammatory cyto-kines. Macrophages treated with SFN exhibited a significant decrease in the levels of S. aureus-induced inflammation, and while the anti-inflammatory activity of SFN has been previously described, the underlying cellular mechanisms involved in this process remains unclear. The use of Nrf2−/− BMDMs also demonstrated that the inhibitory effects of SFN on intracellular bacterial survival and caspase-3/7-dependent apoptosis were Nrf2-independent, but associated with the downregulation of miR-142-5p and miR-146a-5p.
S. aureus is considered a facultative intracellular pathogen, as it has been demonstrated to proliferate extracellularly and intracellularly within various cell types (29). In chronic and recurrent infections, S. aureus may persist several days within the infected macrophages prior to proliferation (30). The present study first validated that the strain of S. aureus used was able to invade and survive within the THP-1-derived macrophages. Upon bacterial recognition by the host innate immune receptors, cascades of highly dynamic signal transduction systems are sequentially activated and amplified, resulting in the proinflammatory response required in the antimicrobial defense mechanism. Therefore, it stands to reason that bacteria have developed, among their numerous immune evasion strategies, elaborative mechanisms to exploit the host innate immune signal transduction pathways (31). The increase in inflammatory cytokines produced by the inflammatory response in response to bacterial infection has been associated with an increase in the rates of bacterial nosocomial infections (32). Bacteria such as S. aureus, Pseudomonas aeruginosa and Acinetobacter spp., incubated with IL-1β, IL-6 and TNF-α, demonstrated cytokine concentration-dependent growth patterns (32). In patients with atopic dermatitis, S. aureus recognition mediated an increase in the production of inflammatory cytokines IL-1β or IFN-γ and promoted the cytokine-dependent growth enhancement of S. aureus, increasing the severity of the skin lesion (33). A recent study suggested that S. aureus-alpha toxin may activate the inflammasome through activation of the acid sphingomye-linase, which resulted in the rapid release of cathepsins and the production of IL-1β and TNF-α in BMDMs (34). The present study demonstrated that S. aureus infection of macrophages triggered a robust increase in the transcriptional expression levels of proinflammatory genes, which was associated with increased expression levels of M1 marker genes. The intracellular survival rates of S. aureus were observed to benefit from this inflammatory environment.
MAPK signaling pathways are central to the inflammatory response of the innate immune system. In human bronchial epithelial cells, concomitant infection of influenza virus and S. aureus synergistically promoted enhanced phosphorylation of p38, ERK and JNK. Treatment of these cells with specific inhibitors of p38 and ERK demonstrated that these signaling pathways were associated with the regulation of IL-6 production (35). Herein, the THP-1-derived macrophages and Nrf2+/− BMDMs infected with S. aureus exhibited increased phosphorylation of p38 and JNK MAPK. The rapid inflammatory response was reflected in the marked increase in IL-1β, IL-6, and TNF-α mRNA expression levels in the S. aureus-infected macrophages. IL-1β, IL-6 and TNF-α are proinflammatory cytokines known to initiate and regulate the immune response and inflammation. The use of specific MAPK inhibitors for p38 and JNK signaling allowed a better undestanding of the role served by each member of the MAPK family. Based on the results, it was determined that the transcriptional expression levels of the IL-1β, IL-6 and TNF-α genes were p38- and JNK-dependent.
In previous years, a number of compounds have been developed to inhibit protein kinases, with several drugs approved for the treatment of cancer. For treatments of neuroinflammatory disorders, several anti-inflammatory compounds specifically targeting the classical MAPKs, such as p38α MAPK inhibitors, were assessed in clinical trials but were rapidly discontinued as a result of poor safety profiles or low long-term efficacy (36). Several natural or chemical compounds have also been previously used to modulate the S. aureus-mediated inflammatory response by acting on intracellular signal transduction pathways. In a mouse model of S. aureus-induced mastitis, mice treated with the natural compound brazilin, present in Caesalpinia sappan heartwood, exhibited a decrease in levels of the S. aureus-induced inflammatory cytokines IL-1β, IL-6 and TNF-α, leading to a decrease in inflammatory-mediated tissue injury. In addition, an inhibition of the S. aureus-induced phosphorylation of p38, JNK and ERK in the brazilin-treated mice was observed (37). Similar regulatory mechanisms of NF-κB and MAPK signaling pathways were observed in S. aureus-infected RAW 264.7-derived macrophages treated with selenium derivatives. The downregulation of NF-κB and MAPK signaling pathways by selenium was suggested to be associated with the decrease in TNF-α, IL-1β and IL-6 transcriptional expression levels and their cytokine release (38). In addition, treatment of an S. aureus-induced peritonitis mouse model with an ephedrine derivative increased the survival rate of the infected mice by decreasing inflammation through the modulation of the PI3K/AKT and p38 signaling pathways (39). Based on these data, we hypothesized that SFN, known for its anti-inflammatory and antioxidant properties, was able to affect the S. aureus-mediated increase in the inflammatory response and decrease bacterial survival in S. aureus-infected macrophages. THP-1-derived macrophages, primary PBMC-derived macrophages or primary BMDM, pretreated with SFN and challenged with S. aureus, exhibited a signifi-cant decrease in bacterial survival 24 h after internalization. In addition, SFN significantly inhibited the S. aureus-mediated activation of p38 and JNK MAPK in macrophages. In concordance with these results, inhibition of p38 and JNK signaling with SB203580 and SP600125 inhibitors resulted in similar beneficial effects on macrophages compared with those observed in the SFN-treated macrophages, such as the significant repression of S. aureus-mediated upregulation of the proinflammatory genes IL-1β, IL-6 and TNF-α, and the subsequent decrease in intracellular survival rate of S. aureus.
As the pathogen-associated molecular patterns of S. aureus are recognized by pattern recognition receptors (PRRs), there is a strong possibility that the anti-inflammatory effect of SFN relies on the disruption of the receptors preventing pathogen recognition. Toll-like receptors (TLR) are PRRs that recognize pathogen patterns or cell debris, and activate downstream TLR signaling. S. aureus binds to membrane protein TLR2, thereby activating a cascade of proinflammatory responses through NF-κB and MAPK signaling pathways. Watanabe et al (40) suggested that the TLR2-mediated activation of JNK decreased the level of superoxide in S. aureus-infected macrophages, resulting in the prolonged intracellular survival of S. aureus in phagosomes. In addition, using LPS-stimulated RAW264.7 macrophages, Youn et al (41) demonstrated that SFN suppressed the TLR4-mediated increase in inflammation by hindering TLR4 oligomerization in a thiol-dependent manner. However, alternative pathogen receptors also participated in S. aureus recognition, as TLR2-deficient mice were still able to produce decreased but significant levels of cytokines in response to S. aureus infection (42). Cytosolic receptors such as nucleotide-binding oligomerization domain (NOD)-like receptors, in particular NOD2, participate in intracellular S. aureus recognition (43). Similar to TLR2, NOD2 mediates cytokine responses through activation of the NF-κB and MAPK signaling pathways. Experiments performed on primary epithelial cells isolated from NOD2-deficient mice and TLR2-deficient mice indicated that staphylococcal peptidoglycan co-localized with NOD2 and TLR2. In both mutants, the levels of IL-1β and IL-6 were decreased by one-half compared with the wild-type cells, suggesting the additive effects of each receptor (44). In addition, it is unlikely that SFN disruption of TLR2 oligomerization affects the phagocytosis process of macrophages, as TLR2-deficient macrophages maintain their phagocytic activity (45), which concurs with the similarities in internalized S. aureus levels between DMSO- and SFN-pretreated macrophages observed in the present study. Based on these data, we hypothesize that SFN decreases the recognition of S. aureus with cell wall components by impeding TLR2 heterodimerization, thereby preventing the downstream activation of NF-κB and MAPK signaling pathways, and inhibiting the production of proinflammatory cytokines. Future experiments are required to determine whether SFN interferes with bacterial invasion, intracellular survival, and/or replication, whether the anti-inflammatory and anti-apoptotic molecular mechanisms activated by SFN are effective against methicillin-resistant S. aureus, and whether SFN affects the phagosomal maturation process in bacteria-infected macrophages.
Using BMDMs isolated from the bone marrow of Nrf2+/− and Nrf2−/− mice, the present study demonstrated that Nrf2 was necessary in promoting S. aureus-mediated caspases-3/7-dependent cell death. Previously, the anti-inflammatory contribution of Nrf2 was hypothesized to be an indirect effect of the upregulation of genes coding for antioxidant enzymes, with the removal of ROS resulting in a decreased inflammation (46). Herein, as the ROS levels remained unaffected by either S. aureus challenge or SFN stimulation in THP-1-derived macrophages, the contribution of ROS in the regulation of inflammation by SFN was excluded. Despite the marked activation of ERK MAPK and Nrf2 signaling pathways by SFN, the results of the present study clearly established that SFN affects intracellular bacterial survival independently of Nrf2. In contrast with the observations of the present study, several studies have demonstrated that the inhibition of microbial survival in macrophages treated with SFN involved the Nrf2 signaling pathway. In HIV infection, SFN did not interfere with viral entry, but has been demonstrated to inhibit HIV infection in macrophages through Nrf2 activation by preventing viral insertion into host chromosomes (15). In addition, we have previously reported that intracellular survival of Mycobacterium abscessus was decreased through SFN-induced Nrf2-mediated cell apoptosis (23).
Recent studies have associated the expression levels of several miRNAs with fundamental cellular processes such as cell apoptosis, phagocytosis, inflammation and macrophage polarization upon bacterial infection (20,47). The present study demonstrated that expression levels of miR-142-5p and miR-146a-5p are negatively regulated by SFN, suggesting their involvement in SFN-mediated decrease of cell apoptosis. miR-146a-5p expression levels are induced by inflammatory stimuli including TNF-α and IL-1β, and exert an anti-inflammatory effect by inhibiting pro-inflammatory genes such as IL-1 receptor associated kinase 1 and TNF receptor associated factor 6 in human adipocytes (48,49). We hypothesize that the lower than expected expression levels of miR-142-5p and miR-146a-5p observed in THP-1-derived macrophages infected by S. aureus may be due to the short incubation time (3 h after infection) in the experiment in the present study. Additional experiments, including increasing the infection time period, or using primary cells such as BMDMs or human PBMCs, will improve the understanding of the effect of SFN-mediated negative regulation of miR-142-5p, miR-146a-5p and other miRNAs on cellular processes such as inflammation, phago-cytosis and macrophage polarization, as well as identify their target mRNAs.
In conclusion, this study proposed a more comprehensive view of the molecular mechanism activated by SFN to promote bacteria clearance. Unlike pharmacological compounds that specifically target protein kinases, the present study demonstrated that due to the pleiotropic effects of SFN, SFN may possibly affect a wide range of molecular targets in S. aureus-activated macrophages. The present results suggested that using SFN to regulate the MAPK-inflammatory response pathway and interfere with S. aureus-induced apoptosis may be a promising therapeutic approach to decrease S. aureus intracellular survival.
Supplementary Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Legs Poix funding from the Chancellerie des Universités de Paris (MB), the National Institute of Health and Medical Research funding (MB and TD), and the University of Versailles-Saint-Quentin-en-Yvel ines funding (MB and TD).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TBD conceived, designed and performed the experiments, analyzed and interpreted data, and wrote the manuscript. MA performed the experiments. SV interpreted data and reviewed the manuscript. MB conceived the study, interpreted data and reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocols were approved by the Institutional Animal Ethics Committee CEEA47 PELVIPHARM of the University of Vers ailles-Saint-Quentin-en-Yvelines (Montigny-le-Bretonneux, France), and the Ministry of Higher Education, Research and Innovation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Arango Duque G and Descoteaux A: Macrophage cytokines: Involvement in immunity and infectious diseases. Front Immunol. 5:4912014. View Article : Google Scholar : PubMed/NCBI | |
|
Arthur JS and Ley SC: Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 13:679–692. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Chen GY and Nunez G: Sterile inflammation: Sensing and reacting to damage. Nat Rev Immunol. 10:826–837. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Iwasaki A and Medzhitov R: Regulation of adaptive immunity by the innate immune system. Science. 327:291–295. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Murray PJ and Wynn TA: Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 11:723–737. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Gomez MI, Lee A, Reddy B, Muir A, Soong G, Pitt A, Cheung A and Prince A: Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med. 10:842–848. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Kujime K, Hashimoto S, Gon Y, Shimizu K and Horie T: p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J Immunol. 164:3222–3228. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
DeLeo FR, Otto M, Kreiswirth BN and Chambers HF: Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 375:1557–1568. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Parker D, Ahn D, Cohen T and Prince A: Innate immune signaling activated by MDR bacteria in the airway. Physiol Rev. 96:19–53. 2016. View Article : Google Scholar : | |
|
Fraunholz M and Sinha B: Intracellular Staphylococcus aureus: Live-in and let die. Front Cell Infect Microbiol. 2:432012. View Article : Google Scholar : PubMed/NCBI | |
|
Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P and Lozniewski A: Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a] pyrene-induced stomach tumors. Proc Natl Acad Sci USA. 99:7610–7615. 2002. View Article : Google Scholar | |
|
Romeo L, Iori R, Rollin P, Bramanti P and Mazzon E: Isothiocyanates: An overview of their antimicrobial activity against human infections. Molecules. 23:E6242018. View Article : Google Scholar : PubMed/NCBI | |
|
Sita G, Hrelia P, Graziosi A and Morroni F: Sulforaphane from cruciferous vegetables: Recent advances to improve glioblastoma treatment. Nutrients. 10:E17552018. View Article : Google Scholar : PubMed/NCBI | |
|
Johansson NL, Pavia CS and Chiao JW: Growth inhibition of a spectrum of bacterial and fungal pathogens by sulforaphane, an isothiocyanate product found in broccoli and other cruciferous vegetables. Planta Med. 74:747–750. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Furuya AK, Sharifi HJ, Jellinger RM, Cristofano P, Shi B and de Noronha CM: Sulforaphane Inhibits HIV Infection of Macrophages through Nrf2. PLoS Pathog. 12:e10055812016. View Article : Google Scholar : PubMed/NCBI | |
|
Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al: An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 236:313–322. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, Kelly V, Sekizawa K, Uchida K and Yamamoto M: Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2). Mol Cell Biol. 24:36–45. 2004. View Article : Google Scholar : | |
|
Ishii Y, Itoh K, Morishima Y, Kimura T, Kiwamoto T, Iizuka T, Hegab AE, Hosoya T, Nomura A, Sakamoto T, et al: Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J Immunol. 175:6968–6975. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K and Yamamoto M: Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 7:116242016. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou X, Li X and Wu M: miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther. 3:142018. View Article : Google Scholar : PubMed/NCBI | |
|
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R and Rajewsky N: Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Fuentes F, Paredes-Gonzalez X and Kong AN: Dietary glucosinolates sulforaphane, phenethyl isothiocyanate, indole-3-carbinol/3,3′-diin-dolylmethane: Anti-oxidative stress/inflammation, Nrf2, epigenetics/epigenomics and in vivo cancer chemopreventive efficacy. Curr Pharmacol Rep. 1:179–196. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Bonay M, Roux AL, Floquet J, Retory Y, Herrmann JL, Lofaso F and Deramaudt TB: Caspase-independent apoptosis in infected macrophages triggered by sulforaphane via Nrf2/p38 signaling pathways. Cell death Discov. 1:150222015. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar | |
|
Jorgensen I, Rayamajhi M and Miao EA: Programmed cell death as a defence against infection. Nat Rev Immunol. 17:151–164. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng D, Li J, Zhang L and Hu L: miR-142-5p suppresses proliferation and promotes apoptosis of human osteosarcoma cell line, HOS, by targeting PLA2G16 through the ERK1/2 signaling pathway. Oncol Lett. 17:1363–1371. 2019.PubMed/NCBI | |
|
Shu L, Zhang W, Huang G, Huang C, Zhu X, Su G and Xu J: Troxerutin attenuates myocardial cell apoptosis following myocardial ischemia-reperfusion injury through inhibition of miR-146a-5p expression. J Cell Physiol. 234:9274–9282. 2019. View Article : Google Scholar | |
|
Atri C, Guerfali FZ and Laouini D: Role of human macrophage polarization in inflammation during infectious diseases. Int J Mol Sci. 19:E18012018. View Article : Google Scholar : PubMed/NCBI | |
|
Sendi P and Proctor RA: Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends Microbiol. 17:54–58. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Hamza T and Li B: Differential responses of osteoblasts and macrophages upon Staphylococcus aureus infection. BMC Microbiol. 14:2072014. View Article : Google Scholar : PubMed/NCBI | |
|
Reddick LE and Alto NM: Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol Cell. 54:321–328. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Meduri GU, Kanangat S, Stefan J, Tolley E and Schaberg D: Cytokines IL-1beta, IL-6, and TNF-alpha enhance in vitro growth of bacteria. Am J Respir Crit Care Med. 160:961–967. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Di Domenico EG, Cavallo I, Bordignon V, Prignano G, Sperduti I, Gurtner A, Trento E, Toma L, Pimpinelli F, Capitanio B and Ensoli F: Inflammatory cytokines and biofilm production sustain Staphylococcus aureus outgrowth and persistence: A pivotal interplay in the pathogenesis of atopic dermatitis. Sci Rep. 8:95732018. View Article : Google Scholar : PubMed/NCBI | |
|
Ma J, Gulbins E, Edwards MJ, Caldwell CC, Fraunholz M and Becker KA: Staphylococcus aureus α-toxin induces inflammatory cytokines via lysosomal acid sphingomyelinase and ceramides. Cell Physiol Biochem. 43:2170–2184. 2017. View Article : Google Scholar | |
|
Klemm C, Bruchhagen C, van Kruchten A, Niemann S, Löffler B, Peters G, Ludwig S and Ehrhardt C: Mitogen-activated protein kinases (MAPKs) regulate IL-6 over-production during concomitant influenza virus and Staphylococcus aureus infection. Sci Rep. 7:424732017. View Article : Google Scholar : PubMed/NCBI | |
|
Cohen P: Targeting protein kinases for the development of anti-inflammatory drugs. Curr Opin Cell Biol. 21:317–324. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Gao XJ, Wang TC, Zhang ZC, Cao YG, Zhang NS and Guo MY: Brazilin plays an anti-inflammatory role with regulating Toll-like receptor 2 and TLR 2 downstream pathways in Staphylococcus aureus-induced mastitis in mice. Int Immunopharmacol. 27:130–137. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Bi CL, Wang H, Wang YJ, Sun J, Dong JS, Meng X and Li JJ: Selenium inhibits Staphylococcus aureus-induced inflammation by suppressing the activation of the NF-κB and MAPK signalling pathways in RAW264.7 macrophages. Eur J Pharmacol. 780:159–165. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
He W, Ma J, Chen Y, Jiang X, Wang Y, Shi T, Zhang Q, Yang Y, Jiang X, Yin S, et al: Ephedrine hydrochloride protects mice from Staphylococcus aureus-induced peritonitis. Am J Transl Res. 10:670–683. 2018. | |
|
Watanabe I, Ichiki M, Shiratsuchi A and Nakanishi Y: TLR2-mediated survival of Staphylococcus aureus in macrophages: A novel bacterial strategy against host innate immunity. J Immunol. 178:4917–4925. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Youn HS, Kim YS, Park ZY, Kim SY, Choi NY, Joung SM, Seo JA, Lim KM, Kwak MK, Hwang DH and Lee JY: Sulforaphane suppresses oligomerization of TLR4 in a thiol-dependent manner. J Immunol. 184:411–419. 2010. View Article : Google Scholar | |
|
Takeuchi O, Hoshino K and Akira S: Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 165:5392–5396. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, Nizet V and Eckmann L: NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci USA. 106:12873–12878. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Muller-Anstett MA, Muller P, Albrecht T, Nega M, Wagener J, Gao Q, Kaesler S, Schaller M, Biedermann T and Götz F: Staphylococcal peptidoglycan co-localizes with Nod2 and TLR2 and activates innate immune response via both receptors in primary murine keratinocytes. PLoS One. 5:e131532010. View Article : Google Scholar : PubMed/NCBI | |
|
Villamon E, Gozalbo D, Roig P, O'Connor JE, Fradelizi D and Gil ML: Toll-like receptor-2 is essential in murine defenses against Candida albicans infections. Microbes infect. 6:1–7. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Kong X, Thimmulappa R, Kombairaju P and Biswal S: NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J Immunol. 185:569–577. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Tanaka K, Kim SE, Yano H, Matsumoto G, Ohuchida R, Ishikura Y, Araki M, Araki K, Park S, Komatsu T, et al: MiR-142 is required for staphylococcus aureus clearance at skin wound sites via small GTPase-mediated regulation of the neutrophil actin cytoskeleton. J Invest Dermatol. 137:931–940. 2017. View Article : Google Scholar | |
|
Lambert KA, Roff AN, Panganiban RP, Douglas S and Ishmael FT: MicroRNA-146a is induced by inflammatory stimuli in airway epithelial cells and augments the anti-inflammatory effects of glucocorticoids. PLoS One. 13:e02054342018. View Article : Google Scholar : PubMed/NCBI | |
|
Roos J, Enlund E, Funcke JB, Tews D, Holzmann K, Debatin KM, Wabitsch M and Fischer-Posovszky P: miR-146a-mediated suppression of the inflammatory response in human adipocytes. Sci Rep. 6:383392016. View Article : Google Scholar : PubMed/NCBI |