Identification of novel molecules and pathways associated with fascin actin‑bundling protein 1 in laryngeal squamous cell carcinoma through comprehensive transcriptome analysis
- Authors:
- Published online on: February 28, 2024 https://doi.org/10.3892/ijmm.2024.5363
- Article Number: 39
-
Copyright: © Liu et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Laryngeal squamous cell carcinoma (LSCC) is a common type of head and neck cancer with an increasing incidence and mortality rate (1,2). In 2020, there were 184,615 new LSCC cases and 99,840 LSCC-related deaths (3). Early-stage LSCC is often curable. However, up to 60% of patients are already in the advanced stages of LSCC (clinical stages III or IV) at the time of diagnosis (2). Despite notable advancements in diagnostic and treatment strategies, the 5-year survival rate of patients with LSCC has declined from 66 to 61% over the past 40 years (4). This decline is mainly attributed to the proneness of LSCC to local invasion, cervical lymph node metastasis and the unclear mechanisms of LSCC tumorigenesis or development. Therefore, understanding the molecular regulatory mechanisms underlying the progression of LSCC is crucial for its diagnosis, treatment and prognosis.
Fascin actin-bundling protein 1 (FSCN1), a globular filamentous actin-binding protein, is abnormally overexpressed in multiple types of human cancer, such as laryngeal (5), bladder (6) and breast cancer (7), and elevated FSCN1 levels have been shown to be a hallmark of an aggressive clinical progression and a poor prognosis (8-10). In vitro functional studies using cancer cell lines have revealed that FSCN1 promotes cell growth, migration, invasion and metastasis in various types of cancer, such as oral cancer (11), osteosarcoma (12) and non-small cell lung cancer (8,13). Immunohistochemical studies have also shown that FSCN1 protein expression is associated with aggressive clinical phenotypes, a poor prognosis and shorter survival outcomes (14,15). In addition, FSCN1 has been established to play a role in cancer cell epithelial-mesenchymal transition (5,16), glycolysis (17) and ferroptosis (18). In a previous study by the authors, it was found that FSCN1 expression was substantially upregulated in LSCC tissues compared with adjacent normal tissues (19). Furthermore, FSCN1 knockdown using specific small interfering RNA (siRNA) has been shown to inhibit LSCC cell migration, invasion and growth (5). Therefore, it is necessary to further investigate the regulatory role of FSCN1 in LSCC and to elucidate the molecular mechanisms through which FSCN1 promotes the progression of LSCC.
In the present study, a comprehensive transcriptome analysis using cells in which FSCN1 was knocked down compared with control FSCN1-expressing cells was performed. It was found that FSCN1 knockdown affected the expression of numerous genes. Through a series of analyses, FSCN1 was linked to multiple biological functions, such as transcriptional regulation, response to radiation, focal adhesion, extracellular matrix (ECM)-receptor interaction and steroid biosynthesis. The crosstalk analysis with FSCN1-interacting proteins and co-immunoprecipitation (co-IP) validation revealed that 24-dehydrocholesterol reductase (DHCR24) is a new molecule associated with FSCN1 in LSCC. Further studies focusing on the DHCR24-FSCN1 interaction may provide valuable insight into the molecular mechanisms underlying LSCC.
Materials and methods
Cells and cell culture
The FD-LSC-1 human LSCC cell line [a gift from Professor Liang Zhou (20)] was cultured in BEGM™ Bronchial Epithelial Cell Growth Medium (Lonza Group Ltd.) supplemented with 10% fetal bovine serum (Biological Industries). 293T cells (China Center for Type Culture Collection) and the TU-177 human LSCC cell line (Shanghai Bioleaf Biotech Co., Ltd.) were cultured in DMEM supplemented with 10% fetal bovine serum. The cultures were maintained at 37°C in 5% CO2.
Plasmid construction and cell transfection
Human prostaglandin reductase 1 (PTGR1), DHCR24 and solute carrier family 38 member 2 (SLC38A2) overexpression plasmids were generated by inserting their respective CDS sequence into the p3xFLAG-CMV-10 vector (Merck KGaA). The FSCN1 overexpression plasmid was generated by inserting the FSCN1 coding sequence into the pCMV-HA vector (Clontech; Takara Bio USA, Inc.). Empty p3xFLAG-CMV-10 vector was used as a negative control. For DHCR24 overexpression, FD-LSC-1 and TU-177 cells were seeded in six-well plates at a density of 5×105 cells/well. A total of 5 μl Lipofectamine 3000® (Invitrogen; Thermo Fisher Scientific, Inc.) and 2.5 μg DHCR24 overexpression vector or empty p3xFLAG-CMV-10 vector were mixed in 250 μl Opti-MEM™ (Gibco; Thermo Fisher Scientific, Inc.) and added to each well. Following 6 h of incubation at 37°C with 5% CO2, the medium was replaced with fresh medium. At 48 h following transfection, the cells were collected for use in subsequent experiments.
Small interfering RNA(RNA)
siRNA targeting FSCN1 (si-FSCN1) and scrambled siRNA (si-NC) were synthesized by GenePharma Co., Ltd. The siRNA sequences were as follows: si-FSCN1 sense 5′-GCAAGAAUGCCAGCUGCUACU-3′ and antisense 5′-AGUAGCAGCUGGCAUUCUUGC-3′; and si-NC sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. For transfection, TU-177 cells were seeded in six-well plates at a density of 5×105 cells/well. A total of 5 μl Lipofectamine 3000® (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 nM siRNAs were mixed in 250 μl Opti-MEM™ (Gibco; Thermo Fisher Scientific, Inc.) and added to each well. Following 6 h of incubation at 37°C with 5% CO2, the medium was replaced with fresh medium. At 48 h following transfection, the cells were collected for use in subsequent experiments.
Microarray assay and data analysis
TU-177 cells in which FSCN1 was knocked down and control TU-177 cells were generated using siRNA transfection, followed by total RNA extraction using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Total RNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.), and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Sample labeling, microarray hybridization and washing were performed according to the manufacturer's instructions (Agilent Human lncRNA Microarray 2018 Version, 4×180k, Design ID: 085630). Briefly, total RNA was transcribed into double-stranded cDNA (RNA Spike In Kit, one-color, Agilent p/n 5188-5282; Agilent Technologies, Inc.), synthesized into cRNA (RNA Spike In Kit, one-color, Agilent p/n 5188-5282; Agilent Technologies, Inc.), and labeled with Cyanine-3-CTP (Low Input Quick-Amp Labeling Kit, one-color. Agilent p/n 5190-2305; Agilent Technologies, Inc.). The labeled cRNAs were hybridized onto the microarray. Following washing, the arrays were scanned using the Agilent Scanner G2505C (Agilent Technologies, Inc.).
Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used to analyze array images to obtain raw data, and GeneSpring (version 13.1, Agilent Technologies) was then used to complete the basic analysis of the raw data. First, the raw data were normalized using the quantile algorithm. The probes that at least 1 out of 2 conditions have flags in 'P' were selected for further data analysis. Differentially expressed genes (DEGs) or long non-coding RNAs (lncRNAs) were identified using fold change (FC) and the P-value from the unpaired t-test. The thresholds for up- and downregulated genes were a fold change of ≥1.5 and P≤0.05. Hierarchical clustering was performed to display the expression patterns of distinguishable genes among samples. The microarray assay data were deposited at the Gene Expression Omnibus (GEO) database and are accessible via the accession no. GSE255143.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes Enrichment (KEGG) analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources (http://david.ncifcrf.gov) was used for functional enrichment analysis of GO terms and KEGG pathways. P<0.05 was selected as the cut-off criterion. The GO terms included biological process, cellular component and molecular function. The top 20 GO terms and KEGG pathways were plotted according to their P-values.
Co-expression analysis
The RNA sequencing data of 57 pairs of LSCC and matched adjacent normal mucosa tissues (GSE127165) were downloaded from the GEO database. Of the 1,063 DEGs in the cells in which FSCN1 was knocked down, 641 were expressed in the 57 LSCC tissues. The co-expression between FSCN1 and the 641 DEGs was then calculated using Pearson's correlation analysis, and Pearson's correlation coefficients of <0.4 or >-0.4 were eliminated. Both up- and downregulated DEGs that were positively and negatively correlated with FSCN1, respectively, were eliminated.
Protein-protein interaction (PPI) network analysis
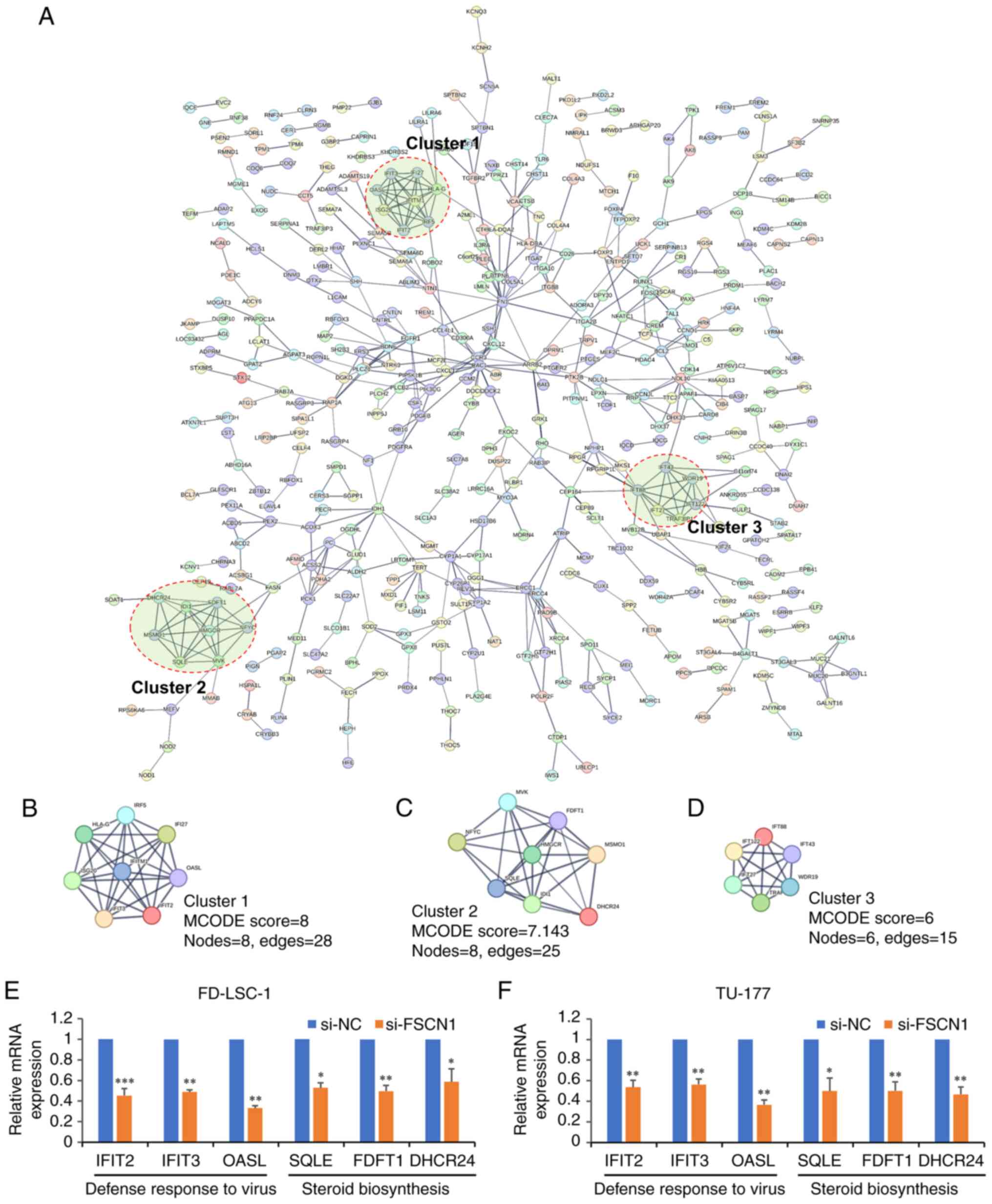
The identified FSCN1-regulated genes were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online tool v11.5 (http://www.string-db.org) for interaction networks. The PPI network was generated with a confidence score of ≥0.7 (high confidence). The highly connected clusters from the PPI network were identified using the 'Molecular Complex Detection (MCODE)' plugin tool in Cytoscape v3.7.1 software. The top three high ranking clusters were selected for further analysis.
Co-IP for western blot analysis
293T cells were cultured in 60-mm plates and each plate was transfected with 5 μg FSCN1 overexpression vector and 5 μg PTGR1 or DHCR24 or SLC38A2 overexpression vector using Lipofectamine 3000® (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 for 6 h, according to the manufacturer's instructions. At 48 h following transfection, the cells were collected for use in subsequent co-IP experiments. Whole-cell extracts were collected by lysing 1×107 cells in Pierce IP Lysis buffer (Thermo Fisher Scientific, Inc.) with the addition of a complete protease inhibitor mixture for 30 min on ice, and clarified using centrifugation at 12,000 x g for 15 min at 4°C. For co-IP, protein extracts were incubated at 4°C overnight with mouse monoclonal anti-flag antibody (1:500; cat. no. F1804; Merck KGaA) or control mouse immunoglobulin G (IgG) (1:500; cat.no. A7028; Beyotime Institute of Biotechnology), and precipitated proteins were captured using Pierce Protein A/G magnetic beads (Thermo Fisher Scientific, Inc.). Following three washes with Pierce IP lysis buffer (Thermo Fisher Scientific, Inc.), bound proteins were eluted in 2X SDS loading buffer and examined using western blot analysis. Rabbit HA tag antibody (1:1,000; cat. no. 51064-2-AP; Proteintech Group, Inc.) was used for western blot analysis.
Western blot analysis
The cells were lysed using Pierce RIPA lysis buffer (Thermo Fisher Scientific, Inc.) containing protease inhibitor cocktail for 30 min on ice. The protein concentration was determined using a BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), and total protein was separated using 10% SDS-PAGE gels, transferred to a PVDF membrane and blocked with 10% w/v non-fat milk powder in TBST at room temperature for 2 h. The membranes were incubated with primary antibodies against Flag (1:1,000; cat. no. F1804; Merck KGaA), FSCN1 (1:1,000; cat. no. 54545S; Cell Signaling Technology, Inc.), and GAPDH (1:1,000; cat.no. HC301-02; TransGen Biotech Co., Ltd.)overnight at 4°C, followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (1:1,000; cat. no. A0216; Beyotime Institute of Biotechnology) at room temperature for 2 h. Immunoreactive bands were visualized using WesternBright® ECL HRP substrate (Advansta Inc.).
Cell proliferation assay
Cell proliferation was determined using the Cell Counting Kit-8 (TransGen Biotech Co., Ltd.), following the manufacturer's instructions. Briefly, 3,000 cells were seeded into each well of a 96-well plate. At 0, 12, 24, 48 and 72 h after seeding, each well was replaced with 100 μl fresh complete medium and 10 μl CCK-8 (TransGen Biotech Co., Ltd.) followed by incubation at 37°C with 5% CO2 for 1 h. The absorbance of the solution was measured at 450 nm using a Spectra Max i3x Multifunctional microplate detection system (Molecular Devices, LLC). A total of three independent experiments were performed.
Cell migration assay
Following transfection, the cells were suspended in a serum-free medium. Serum-free DMEM (200 μl) containing 1×105 cells were added to the upper chamber. A total of 500 μl DMEM medium supplemented with 20% fetal bovine serum (Biological Industries) was then added to the lower chamber. At 48 h following incubation in 37°C with 5% CO2, the cells in the upper chamber were removed with cotton swabs and the lower side of the chamber was gently washed twice with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde for 20 min at room temperature. The cells stained with 0.1% crystal violet (Amresco, LLC) for 10 min at room temperature, and images were then captured using an inverted microscope (Leica Microsystems GmbH).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was isolated from the cells in which FSCN1 was knocked down or control cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. For RT-qPCR of messenger RNA (mRNA), cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis kit (Vazyme Biotech Co., Ltd. according to the following conditions: 25°C for 5 min, 50°C for 15 min, and 85°C for 2 min. ChamQ SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.) was used for qPCR. The following thermocycling conditions were used: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 10 sec. The relative expression level of target genes was calculated using the 2(−ΔΔCq) method (21). The primer sequences used are listed in Table SI.
Statistical analysis
Statistical analysis was performed using SPSS software v22.0 (IBM Corp.). The data were analyzed using the two-tailed Student's t-test; P<0.05 was considered to indicate a statistically significant difference.
Results
Screening of DEGs in LSCC TU-177 cells in which FSCN1 was knocked down
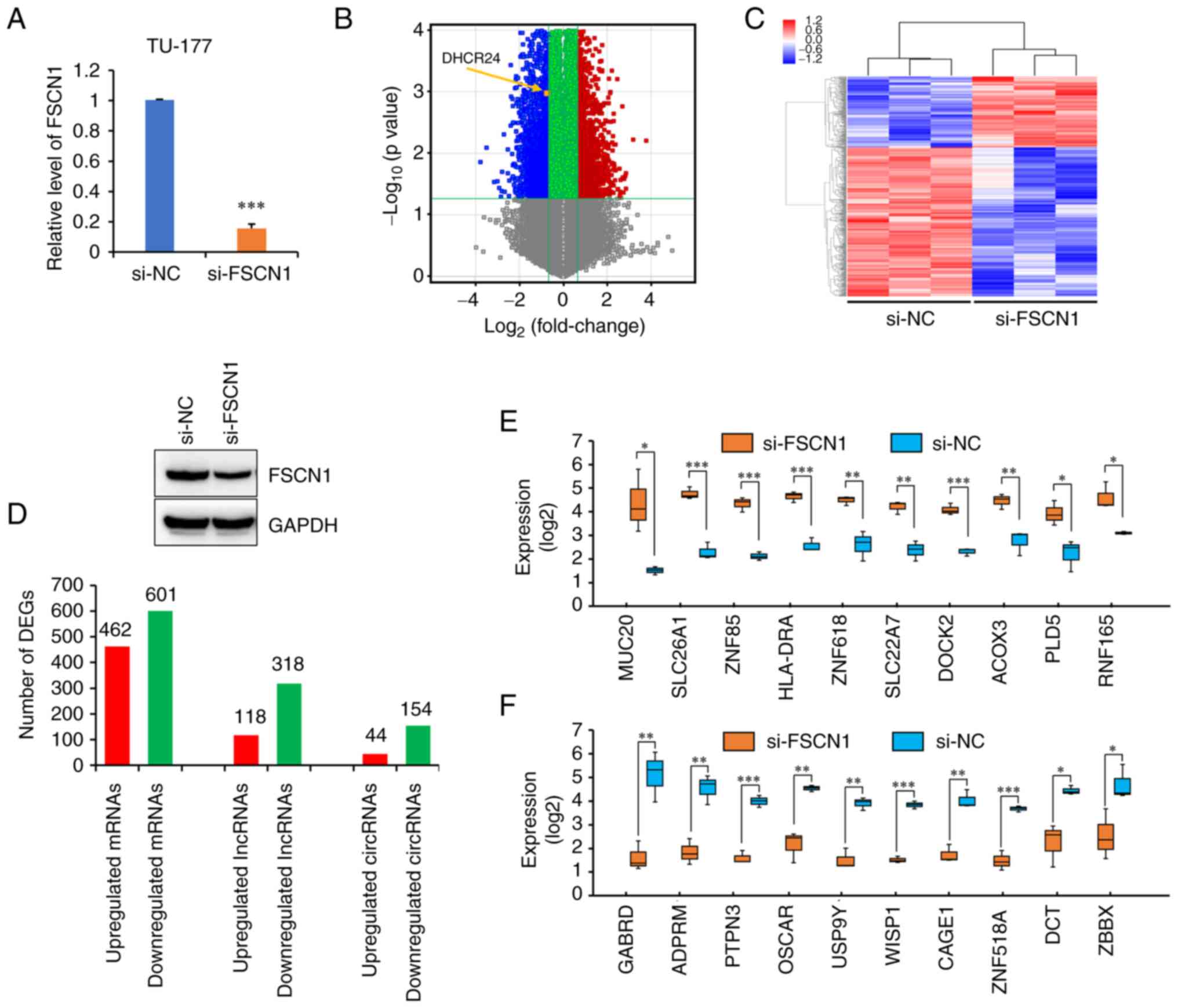
To investigate genes regulated by FSCN1, the microarray analysis of differentially up- or downregulated genes was performed by knocking down FSCN1 using siRNAs in TU-177 cells. The results of RT-qPCR indicated that FSCN1 was successfully knocked down (Fig. 1A). Following data preprocessing, 1,063 DEGs (462 up- and 601 downregulated) were identified in the TU-177 cells transfected with FSCN1 siRNAs compared with those transfected with the control under the thresholds of FC>1.5 and P<0.05. Based on the volcano plot and hierarchical clustering analysis of these DEGs, the control and FSCN1 knockdown groups were clearly distinguished (Fig. 1B and C). In addition, 436 differentially expressed lncRNAs (118 up- and 318 downregulated) and 198 differentially expressed circular RNAs (circRNAs) (44 up- and 154 downregulated) were obtained between the FSCN1 knockdown and control groups (Fig. 1D). These genes are listed in Table SII. In addition, the expression of the top 10 up- and downregulated genes in TU-177 cells in which FSCN1 was knocked down compared with the control cells based on microarray data are shown in Fig. 1E and F.
Function annotation of differentially expressed mRNAs
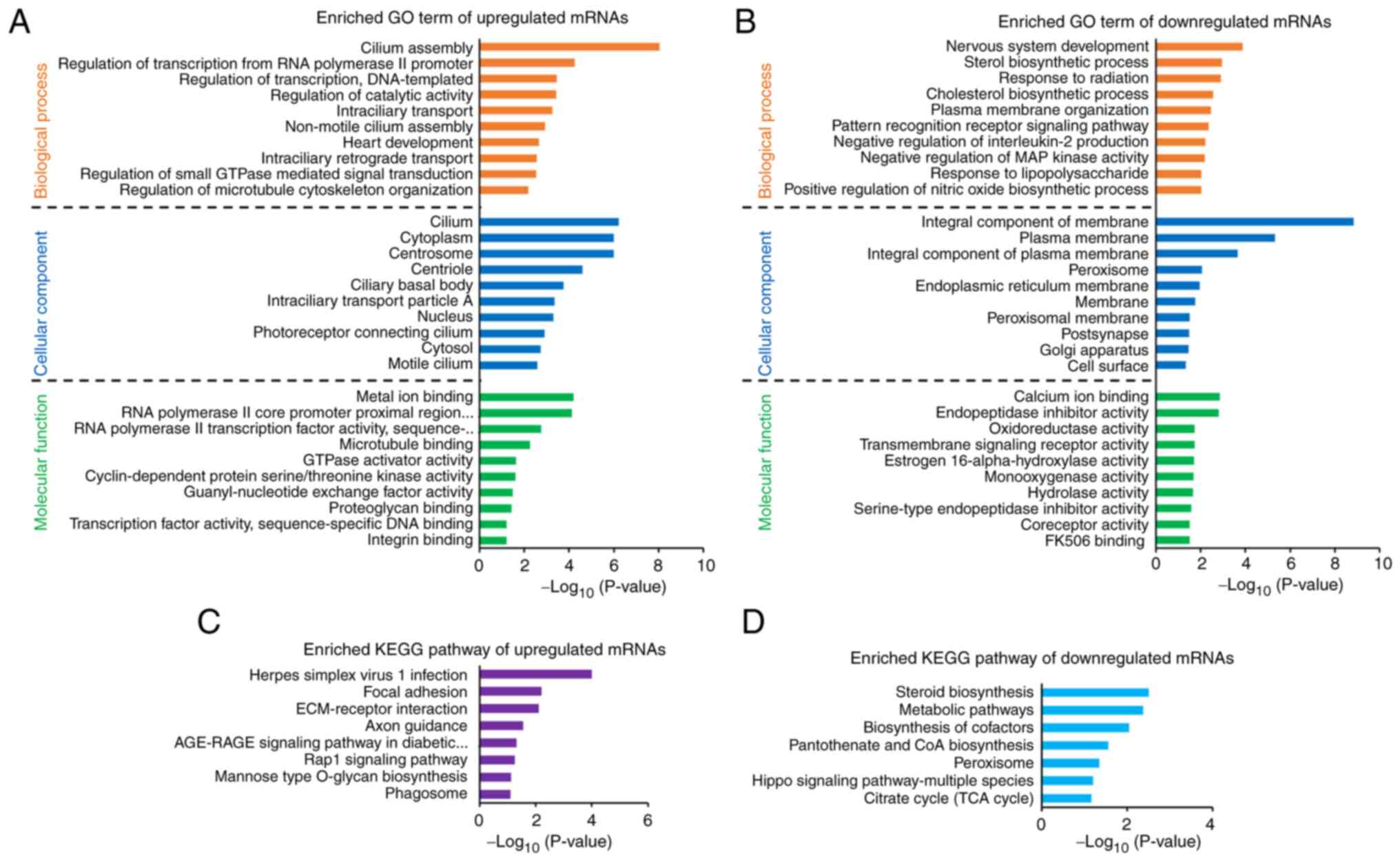
To identify the putative functions and pathways associated with the FSCN1-regulated genes, functional enrichment analysis was performed through the DAVID Bioinformatics Resources (22). The upregulated DEGs were significantly enriched with biological functions of 'cilium assembly', 'regulation of transcription from RNA polymerase II promoter' and 'regulation of transcription, DNA-templated' (Fig. 2A). By contrast, the downregulated DEGs were associated with 'nervous system development', 'sterol biosynthetic process' and 'response to radiation' (Fig. 2B). In the cellular components category, 'cilium' and 'cytoplasm' were significantly enriched using the upregulated DEGs, whereas 'integral component of membrane' and 'plasma membrane' were involved with the downregulated DEGs (Fig. 2A and B). For the GO molecular functions category, the upregulated DEGs were enriched with functions, such as 'metal ion binding', 'RNA polymerase II core promoter proximal region sequence-specific DNA binding' and 'RNA polymerase II transcription factor activity, sequence-specific DNA binding' (Fig. 2A). The downregulated DEGs were associated with 'calcium ion binding', 'endopeptidase inhibitor activity' and 'oxidoreductase activity' (Fig. 2B). According to the findings of KEGG pathway analysis, the upregulated DEGs demonstrated significant enrichment in the pathways of 'herpes simplex virus 1 infection', 'focal adhesion' and 'ECM-receptor interaction' (Fig. 2C). By contrast, the downregulated DEGs were involved in 'steroid biosynthesis', 'metabolic pathways' and 'biosynthesis of cofactors' (Fig. 2D). The complete results are presented in Table SIII.
Construction of a network of genes co-expressed with FSCN1 in LSCC
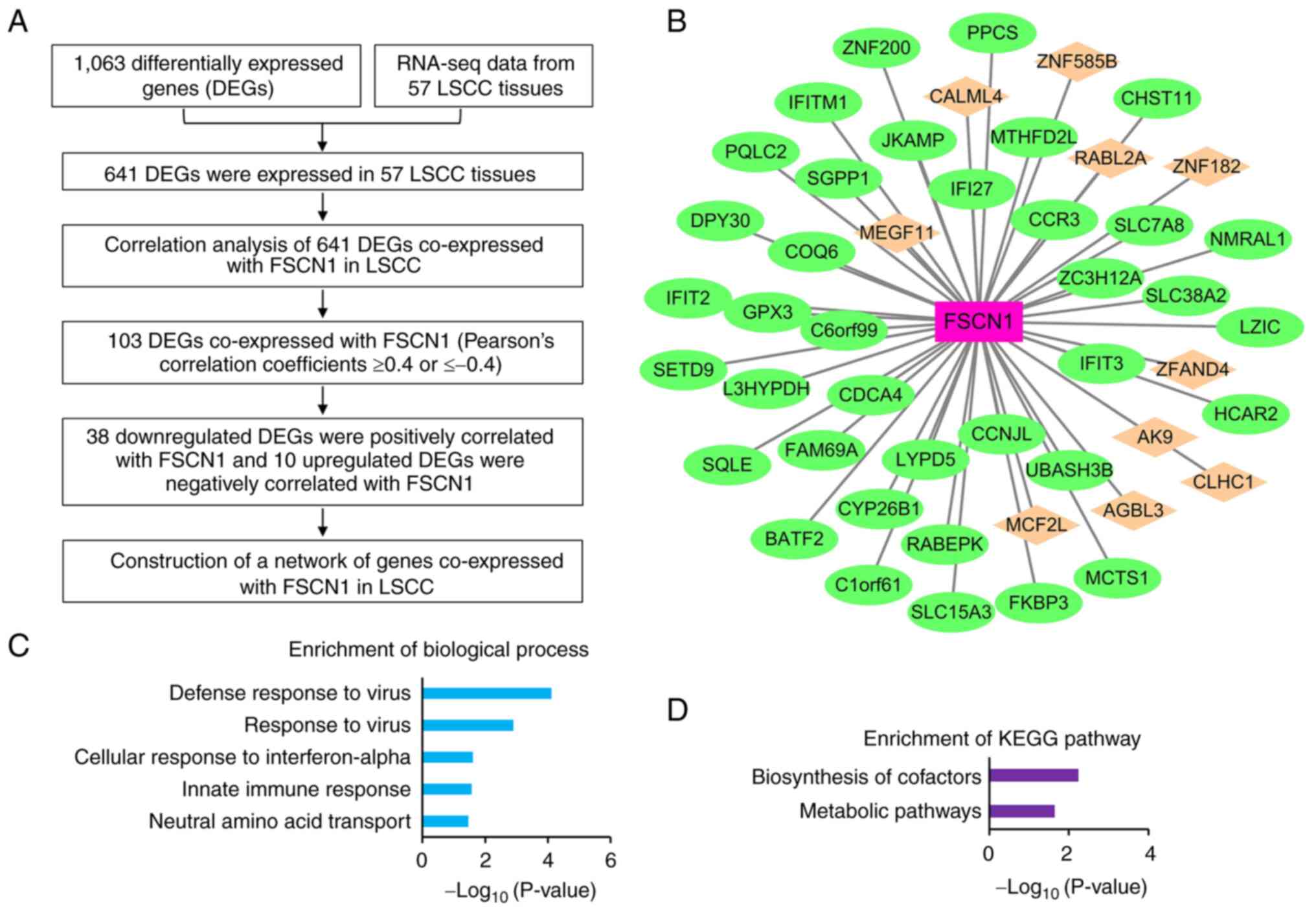
RNA sequencing (RNA-seq) was previously performed in 57 pairs of LSCC and matched adjacent normal mucosa tissues to construct mRNA expression profiles (23). Based on the aforementioned RNA-seq data and 1,063 DEGs in the cells in which FSCN1 was knocked down, a network of genes co-expressed with FSCN1 in LSCC was constructed using the flowchart illustrated in Fig. 3A. Finally, 48 DEGs (10 up- and 38 downregulated DEGs) were found to be co-expressed with FSCN1 in LSCC with Pearson's correlation coefficients of 0.4. A network was constructed and visualized using Cytoscape software (Fig. 3B). In addition, functional enrichment analysis revealed that the 48 genes co-expressed with FSCN1 were significantly associated with the functions of 'defense response to virus', 'response to virus', 'cellular response to interferon-α', 'innate immune response' and 'neutral amino acid transport' (Fig. 3C). KEGG pathway analysis demonstrated that 'biosynthesis pathways of cofactors' and 'metabolic pathways' were significantly enriched by genes co-expressed with FSCN1 (Fig. 3D). The complete results are presented in Table SIV.
PPI network of DEGs
To explore the interactions among these FSCN1-regulated genes and identify specific functional complexes, a PPI network was constructed using the STRING database. In the PPI network, the minimum required interaction score was set as >0.700 (high confidence), and disconnected nodes in the network were hidden. A PPI network containing 455 nodes and 570 edges was constructed (Fig. 4A). In addition, three highly connected clusters were identified from the PPI network using the MCODE plugin tool (24) in Cytoscape (Fig. 4B-D). The GO and KEGG enrichment analyses revealed that Cluster 1 was mainly involved with functions of defense response to virus (GO:0051607) and response to virus (GO:0009615). By contrast, Cluster 2 was associated with the function of cholesterol biosynthetic process (GO:0006695) and the KEGG pathway of steroid biosynthesis (hsa00100). The genes in Cluster 3 were mainly associated with cilium assembly (GO:0060271).
Subsequently, RT-qPCR was used to validate the expression of genes involved in the function of defense response to virus and steroid biosynthesis pathway. Compared with the control group, interferon induced protein with tetratricopeptide repeats (IFIT)2, IFIT3, 2′-5′-oligoadenylate synthetase like (defense response to virus; QASL), squalene epoxidase (SQLE), farnesyl-diphosphate farnesyltransferase 1 (FDFT1) and DHCR24 (steroid biosynthesis) were significantly downregulated in the LSCC cells in which FSCN1 was knocked down (Fig. 4E and F), indicating that FSCN1 affected these functions and pathways.
FSCN1-interacting proteins in LSCC and crosstalk analysis
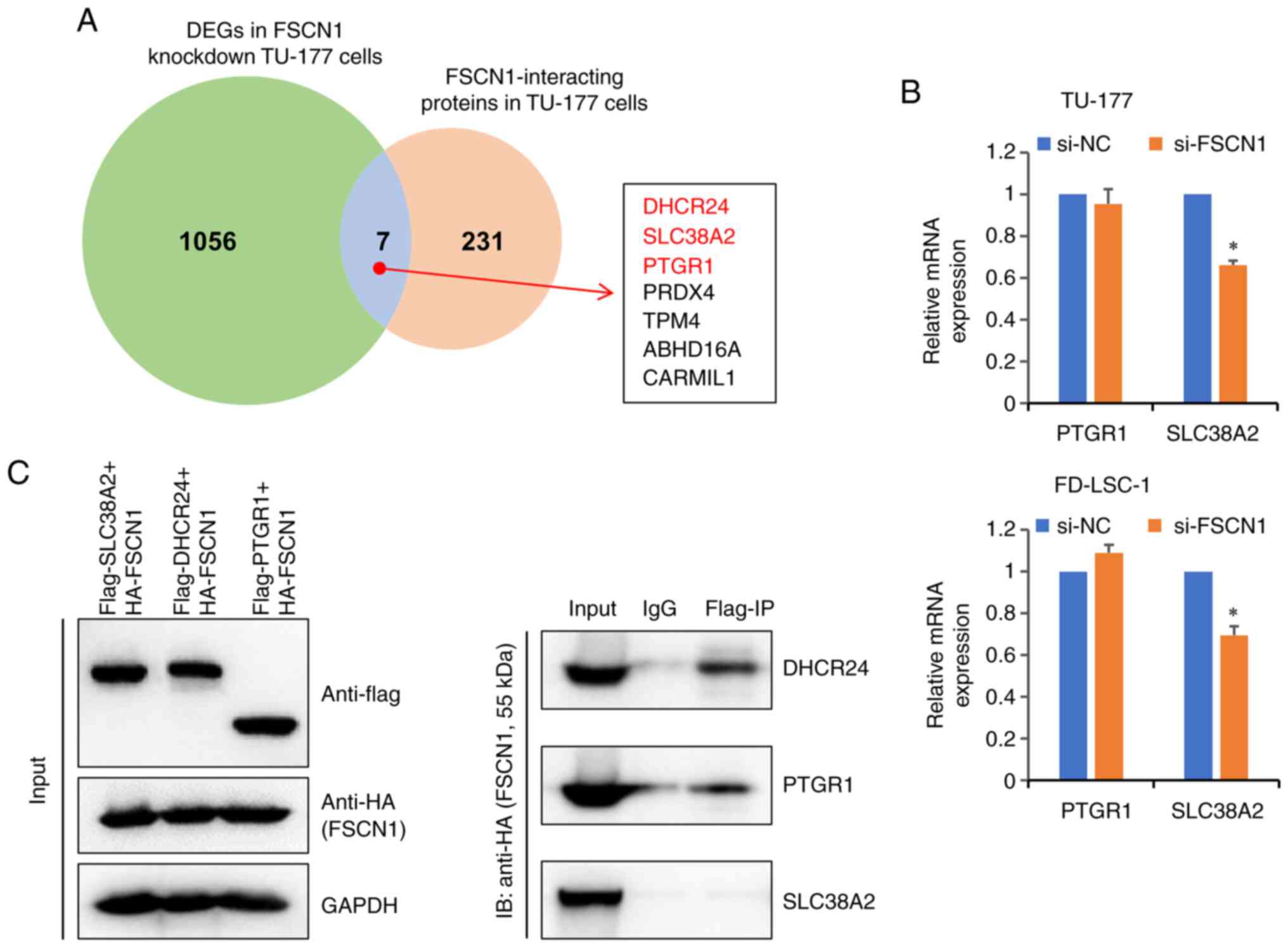
FSCN1-interacting proteins were previously characterized in LSCC cell lines using a mass spectrometry-based proteomics approach, and 238 proteins were identified as interacting partners of FSCN1 in TU-177 cells (25). Following Venn analysis, seven overlapped target genes [DHCR24, SLC38A2, PTGR1, peroxiredoxin 4 (PRDX4), tropomyosin 4 (TMP4), abhydrolase domain containing 16A (ABHD16A), phospholipase and capping protein regulator and myosin 1 linker 1 (CARMIL1)] were identified (Fig. 5A). Of note, it was found that a proven FSCN1-regulated gene (DHCR24; Fig. 4E and F), interacted with FSCN1 in TU-177 cells. SLC38A2, a gene co-expressed with FSCN1 in LSCC (Fig. 3B) interacted with FSCN1 in TU-177 cells. Furthermore, another DEG (PTGR1) in cells in which FSCN1 was knocked down was identified as an FSCN1-interacting protein in Hep-2 cells (25). RT-qPCR was performed to verify the relative expression levels of PTGR1 and SLC38A2 in LSCC cells in which FSCN1 was knocked down. As shown in Fig. 5B, SLC38A2 was significantly downregulated in LSCC cells in which FSCN1 was knocked down, whereas the expression of PTGR1 was not significantly altered.
The present study then attempted to validate the interaction between FSCN1, and PTGR1, DHCR24 and SLC38A2 through co-IP. The three genes were cloned into vectors with a 3xFlag tag, and the FSCN1 gene was cloned into vectors with an HA tag. FSCN1 and Flag-tagged proteins were transiently co-expressed in 293T cells. The expression of all three Flag-tagged proteins and HA-tagged FSCN1 was successfully confirmed using western blot analysis (Fig. 5C). Co-IP was performed on whole cell extracts with Flag-tag antibodies, with matched normal IgG used as a negative control. Out of these three proteins, two were confirmed to bind to FSCN1 (Fig. 5C), and one protein (SLC38A2) did not interact with FSCN1.
Overexpression of DHCR24 promotes the proliferation and migration of LSCC cells
As DHCR24 binds to the FSCN1 protein and its expression is regulated by FSCN1, the expression levels of DHCR24 in the FD-LSC-1 and TU-177 LSCC cell lines were first investigated. Compared with the normal cell line (293T), DHCR24 was significantly downregulated in the FD-LSC-1 and TU-177 LSCC cell lines (Fig. 6A). To further investigate the functional role of DHCR24 in LSCC cells, the DHCR24 overexpression vector (OE-DHCR24) and its empty vector (OE-NC) were transfected into FD-LSC-1 and TU-177 cells. The efficiency of DHCR24 protein overexpression was further verified using western blot analysis (Fig. 6B). Compared with the negative control group, DHCR24 overexpression promoted LSCC cell proliferation (Fig. 6C and D). Furthermore, the results of Transwell assay demonstrated that DHCR24 overexpression markedly promoted LSCC cell migration (Fig. 6E). These findings suggested that DHCR24 promotes the proliferation and migration of LSCC cells.
Discussion
FSCN1 has been widely reported to be overexpressed in several types of human cancer, such as laryngeal (5), bladder (6) and breast cancer (7), and its expression has been associated with an aggressive clinical course, a poor prognosis and shorter survival rates (8,10,26). Previous studies have indicated that FSCN1 expression levels in LSCC tissues are significantly higher than those in adjacent normal tissues, and that FSCN1 knockdown inhibits cell growth, migration and invasion in LSCC (5,19). In addition, a high expression of FSCN1 has been shown to be significantly associated with clinical features and poor outcomes in LSCC (5,19,27,28). However, the regulatory mechanisms of FSCN1 in LSCC remain unclear. In the present study, to gain more insight into the function of FSCN1 in LSCC progression, microarray analysis was performed to screen the DEGs in TU-177 LSCC cells in which FSCN1 was knocked down. Following data preprocessing, 462 genes were upregulated and 601 genes were downregulated in the FSCN1-knockdown cells relative to the control cells with a 1.5-FC cut-off value. According to functional enrichment analysis, these FSCN1-regulated genes were associated with gene transcription (29-31), radioresistance (32) and focal adhesion (33,34), which was consistent with the results of previous studies. Of note, to the best of our knowledge, there is no evidence to date that FSCN1 plays a role in the sterol biosynthetic process, ECM-receptor interaction and the steroid biosynthesis pathway, suggesting that FSCN1 may be linked to previously unknown functions.
To further understand the functional association between these DEGs in TU-177 cells in which FSCN1 was knocked down and FSCN1 in LSCC, a network of genes co-expressed with FSCN1 in LSCC was constructed. The results revealed that 10 up- and 38 downregulated DEGs were found to be co-expressed with FSCN1 in LSCC tissues. Furthermore, functional enrichment analysis and RT-qPCR validation revealed that numerous genes co-expressed with FSCN1 were associated with the functions of the defense response to virus and response to virus, indicating that FSCN1 may play a role in the defense response to a virus. This result was also consistent with the results of the PPI analysis.
Studies have reported that dysregulated cholesterol metabolism is exhibited in cancer cells, with cholesterol and its metabolites acting as signaling molecules that drive tumor development (35,36). Although FSCN1 plays a role in cancer progression, there is no evidence to date to indicate that FSCN1 is associated with cholesterol metabolism. Herein, the results of PPI analysis revealed another highly connected cluster (Cluster 2, Fig. 4C), in which several FSCN1-regulated genes were associated with the cholesterol biosynthetic process and steroid biosynthesis pathway. For example, SQLE is a key rate-limiting enzyme that catalyzes the conversion of squalene to 2,3-epoxysqualene in cholesterol biosynthesis (37,38). FDFT1 is an upstream enzyme of SQLE in cholesterol biosynthesis and plays a key regulatory role in catalyzing the dimerization of two molecules of farnesyl diphosphate to produce squalene (39). DHCR24 is the final enzyme in cholesterol biosynthesis that catalyzes the reduction of the delta-24 double bond in sterol intermediates to form cholesterol (40). To address whether FSCN1 affects cholesterol metabolism, the expression of the aforementioned three relevant genes was validated using RT-qPCR. The expression levels of SQLE, FDFT1 and DHCR24 were significantly downregulated in the LSCC cells in which FSCN1 was knocked down (Fig. 4E and F), suggesting that FSCN1 plays a role in cholesterol synthetic metabolism. Studies have reported that these three genes are involved in cancer development and progression (41-44). Therefore, FSCN1 may regulate LSCC progression by maintaining intracellular cholesterol metabolic homeostasis.
Furthermore, through a crosstalk analysis with FSCN1-interacting proteins in LSCC cells, three genes/proteins (PTGR1, DHCR24 and SLC38A2) were shown to regulate LSCC progression by FSCN1. Following co-IP validation, it was confirmed that DHCR24 and PTGR1 could bind to FSCN1. The interaction between FSCN1 and DHCR24 further strengthened the role of FSCN1 in cholesterol metabolism.
Previous studies have reported that DHCR24 is dysregulated in various types of cancer (41,42,45-47). Its high expression has been shown to be associated with aggressiveness and disease recurrence in endometrial, urothelial and hepatocellular carcinoma (41,42,47). Consistently, the data of the present study revealed that the overexpression of DHCR24 promoted LSCC cell proliferation and migration.
Finally, several studies have been conducted on the potential mechanisms through which FSCN1 affects the expression of a large number of genes (29,31). For example, Saad et al (31) revealed that phosphorylated FSCN1 (p-FSCN1) localized in the nucleus and regulated histone methylation and gene transcription. Mechanistically, p-FSCN1 was shown to specifically interact with the H3K4 methyltransferase core subunit RbBP5 form H3K4me3. Nuclear pFSCN1 interactions with the RNA polymerase II complex further elucidate the role of FSCN1 in gene transcription (31). In addition, FSCN1 knockdown has been reported to affect the expression of a number of genes in various types of cancer cells, such as esophageal squamous cell carcinoma, triple-negative breast cancer and cervical cancer cells (29,30,48). These results indicate that FSCN1 may be a transcriptional regulator.
In conclusion, the present study demonstrated that FSCN1 knockdown affected the expression of up to 1,063 genes, with 462 upregulated and 601 downregulated genes in TU-177 cells. Through crosstalk analysis and validation, FSCN1 was shown to be linked to novel functions, including tje defense response to virus and steroid biosynthesis. In particular, it was found that DHCR24, a key enzyme in cholesterol biosynthesis, interacted with FSCN1, suggesting that FSCN1 may promote LSCC progression by mediating cholesterol metabolism-related signaling pathways. Overall, the present study provided a comprehensive understanding of the diverse functions of FSCN1 and found that the FSCN1-DHCR24 interaction may play a key role in LSCC progression.
Supplementary Data
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HL and WH designed the study and wrote the first draft of the manuscript. CZ and HL revised the manuscript. WH, XW and YZ conducted the bioinformatics analysis. HL, XX and LH developed the methods and performed the validation. LH, XW and XX participated in data analysis and tabulation. YZ, JY and CZ performed the statistical analysis. HL and CZ confirmed the authenticity of all the raw data. All authors have read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81802793), the Basic Research Program of Shanxi Province (Free Exploration) (grant nos. 202203021211015, 20210302124704, 20210302124594 and 202103021223430), the Science Research Start-up Fund for Doctor of Shanxi Medical University (XD1801), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (STIP; grants 2021-185), and the Medical Science and Technology Innovation Team of Shanxi Province (grant no. 2020TD26).
References
|
Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE and Grandis JR: Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 6:922020. View Article : Google Scholar : PubMed/NCBI | |
|
Steuer CE, El-Deiry M, Parks JR, Higgins KA and Saba NF: An update on larynx cancer. CA Cancer J Clin. 67:31–50. 2017. View Article : Google Scholar | |
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Siegel RL, Miller KD, Wagle NS and Jemal A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Gao W, Zhang C, Li W, Li H, Sang J, Zhao Q, Bo Y, Luo H, Zheng X, Lu Y, et al: Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther. 27:365–379. 2019. View Article : Google Scholar : | |
|
Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N and Nakagawa M: miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Ghebeh H, Al-Khaldi S, Olabi S, Al-Dhfyan A, Al-Mohanna F, Barnawi R, Tulbah A, Al-Tweigeri T, Ajarim D and Al-Alwan M: Fascin is involved in the chemotherapeutic resistance of breast cancer cells predominantly via the PI3K/Akt pathway. Br J Cancer. 111:1552–1561. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Zhang Y, Li L, Cao J, Guo Y, Wu Y and Gao W: Fascin actin-bundling protein 1 in human cancer: Promising biomarker or therapeutic target? Mol Ther Oncolytics. 20:240–264. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Li CH, Chan MH, Liang SM, Chang YC and Hsiao M: Fascin-1: Updated biological functions and therapeutic implications in cancer biology. BBA Adv. 2:1000522022. View Article : Google Scholar | |
|
Li Z, Shi J, Zhang N, Zheng X, Jin Y, Wen S, Hu W, Wu Y and Gao W: FSCN1 acts as a promising therapeutic target in the blockade of tumor cell motility: A review of its function, mechanism, and clinical significance. J Cancer. 13:2528–2539. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Alam H, Bhate AV, Gangadaran P, Sawant SS, Salot S, Sehgal L, Dange PP, Chaukar DA, D'cruz AK, Kannanl S, et al: Fascin overexpression promotes neoplastic progression in oral squamous cell carcinoma. BMC Cancer. 12:322012. View Article : Google Scholar : PubMed/NCBI | |
|
Arlt MJ, Kuzmanov A, Snedeker JG, Fuchs B, Silvan U and Sabile AA: Fascin-1 enhances experimental osteosarcoma tumor formation and metastasis and is related to poor patient outcome. BMC Cancer. 19:832019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao J, Zhou Y, Zhang Z, Tian F, Ma N, Liu T, Gu Z and Wang Y: Upregulated fascin1 in non-small cell lung cancer promotes the migration and invasiveness, but not proliferation. Cancer Lett. 290:238–247. 2010. View Article : Google Scholar | |
|
Adams JC: Fascin-1 as a biomarker and prospective therapeutic target in colorectal cancer. Expert Rev Mol Diagn. 15:41–48. 2015. View Article : Google Scholar | |
|
Tan VY, Lewis SJ, Adams JC and Martin RM: Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: A systematic review and meta-analysis. BMC Med. 11:522013. View Article : Google Scholar : PubMed/NCBI | |
|
Wang L, Jia Y, Jiang Z, Gao W and Wang B: FSCN1 is upregulated by SNAI2 and promotes epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Cell Biol Int. 41:833–841. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Lin S, Li Y, Wang D, Huang C, Marino D, Bollt O, Wu C, Taylor MD, Li W, DeNicola GM, et al: Fascin promotes lung cancer growth and metastasis by enhancing glycolysis and PFKFB3 expression. Cancer Lett. 518:230–242. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Chen C, Xie B, Li Z, Chen L, Chen Y, Zhou J, Ju S, Zhou Y, Zhang X, Zhuo W, et al: Fascin enhances the vulnerability of breast cancer to erastin-induced ferroptosis. Cell Death Dis. 13:1502022. View Article : Google Scholar : PubMed/NCBI | |
|
Gao W, Zhang C, Feng Y, Chen G, Wen S, Huangfu H and Wang B: Fascin-1, ezrin and paxillin contribute to the malignant progression and are predictors of clinical prognosis in laryngeal squamous cell carcinoma. PLoS One. 7:e507102012. View Article : Google Scholar : PubMed/NCBI | |
|
Wu CP, Zhou L, Gong HL, Du HD, Tian J, Sun S and Li JY: Establishment and characterization of a novel HPV-negative laryngeal squamous cell carcinoma cell line, FD-LSC-1, with missense and nonsense mutations of TP53 in the DNA-binding domain. Cancer Lett. 342:92–103. 2014. View Article : Google Scholar | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar | |
|
Huang da W, Sherman BT and Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, Niu M, Guo H, Li W, Xue X, et al: Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol Cancer. 19:992020. View Article : Google Scholar : PubMed/NCBI | |
|
Bader GD and Hogue CW: An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Cui J, Zhang Y, Niu M, Xue X, Yin H, Tang Y, Dai L, Dai F, Guo Y, et al: Mass spectrometry-based proteomic analysis of FSCN1-interacting proteins in laryngeal squamous cell carcinoma cells. IUBMB Life. 71:1771–1784. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ristic B, Kopel J, Sherazi S, Gupta S, Sachdeva S, Bansal P, Ali A, Perisetti A and Goyal H: Emerging role of fascin-1 in the pathogenesis, diagnosis, and treatment of the gastrointestinal cancers. Cancers (Basel). 13:25362021. View Article : Google Scholar : PubMed/NCBI | |
|
Zou J, Yang H, Chen F, Zhao H, Lin P, Zhang J, Ye H, Wang L and Liu S: Prognostic significance of fascin-1 and E-cadherin expression in laryngeal squamous cell carcinoma. Eur J Cancer Prev. 19:11–17. 2010. View Article : Google Scholar | |
|
Durmaz A, Kurt B, Ongoru O, Karahatay S, Gerek M and Yalcin S: Significance of fascin expression in laryngeal squamous cell carcinoma. J Laryngol Otol. 124:194–198. 2010. View Article : Google Scholar | |
|
Guo F, Liu Y, Cheng Y, Zhang Q, Quan W, Wei Y and Hong L: Transcriptome analysis reveals the potential biological function of FSCN1 in HeLa cervical cancer cells. PeerJ. 10:e129092022. View Article : Google Scholar : PubMed/NCBI | |
|
Barnawi R, Al-Khaldi S, Majid S, Qattan A, Bakheet T, Fallatah M, Ghebeh H, Alajez NM and Al-Alwan M: Comprehensive transcriptome and pathway analyses revealed central role for fascin in promoting triple-negative breast cancer progression. Pharmaceuticals (Basel). 14:12282021. View Article : Google Scholar : PubMed/NCBI | |
|
Saad A, Bijian K, Qiu D, da Silva SD, Marques M, Chang CH, Nassour H, Ramotar D, Damaraju S, Mackey J, et al: Insights into a novel nuclear function for Fascin in the regulation of the amino-acid transporter SLC3A2. Sci Rep. 6:366992016. View Article : Google Scholar : PubMed/NCBI | |
|
Li S, Huang XT, Wang MY, Chen DP, Li MY, Zhu YY, Yu Y, Zheng L, Qi B and Liu JQ: FSCN1 promotes radiation resistance in patients with PIK3CA gene alteration. Front Oncol. 11:6530052021. View Article : Google Scholar : PubMed/NCBI | |
|
Elkhatib N, Neu MB, Zensen C, Schmoller KM, Louvard D, Bausch AR, Betz T and Vignjevic DM: Fascin plays a role in stress fiber organization and focal adhesion disassembly. Curr Biol. 24:1492–1499. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Villari G, Jayo A, Zanet J, Fitch B, Serrels B, Frame M, Stramer BM, Goult BT and Parsons M: A direct interaction between fascin and microtubules contributes to adhesion dynamics and cell migration. J Cell Sci. 128:4601–4614. 2015.PubMed/NCBI | |
|
Ediriweera MK: Use of cholesterol metabolism for anti-cancer strategies. Drug Discov Today. 27:1033472022. View Article : Google Scholar : PubMed/NCBI | |
|
Xu H, Zhou S, Tang Q, Xia H and Bi F: Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 1874:1883942020. View Article : Google Scholar : PubMed/NCBI | |
|
Gill S, Stevenson J, Kristiana I and Brown AJ: Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13:260–273. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
You W, Ke J, Chen Y, Cai Z, Huang ZP, Hu P and Wu X: SQLE, a key enzyme in cholesterol metabolism, correlates with tumor immune infiltration and immunotherapy outcome of pancreatic adenocarcinoma. Front Immunol. 13:8642442022. View Article : Google Scholar : PubMed/NCBI | |
|
Coman D, Vissers LELM, Riley LG, Kwint MP, Hauck R, Koster J, Geuer S, Hopkins S, Hallinan B, Sweetman L, et al: Squalene synthase deficiency: Clinical, biochemical, and molecular characterization of a defect in cholesterol biosynthesis. Am J Hum Genet. 103:125–130. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Luu W, Zerenturk EJ, Kristiana I, Bucknall MP, Sharpe LJ and Brown AJ: Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis. J Lipid Res. 55:410–420. 2014. View Article : Google Scholar : | |
|
Dai M, Zhu XL, Liu F, Xu QY, Ge QL, Jiang SH, Yang XM, Li J, Wang YH, Wu QK, et al: Cholesterol synthetase DHCR24 induced by insulin aggravates cancer invasion and progesterone resistance in endometrial carcinoma. Sci Rep. 7:414042017. View Article : Google Scholar : PubMed/NCBI | |
|
Lee GT, Ha YS, Jung YS, Moon SK, Kang HW, Lee OJ, Joung JY, Choi YH, Yun SJ, Kim WJ and Kim IY: DHCR24 is an independent predictor of progression in patients with non-muscle-invasive urothelial carcinoma, and its functional role is involved in the aggressive properties of urothelial carcinoma cells. Ann Surg Oncol. 21(Suppl 4): S538–S545. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang H, Tang E, Chen Y, Liu H, Zhao Y, Lin M and He L: Squalene synthase predicts poor prognosis in stage I-III colon adenocarcinoma and synergizes squalene epoxidase to promote tumor progression. Cancer Sci. 113:971–785. 2022. View Article : Google Scholar : | |
|
Wang S, Dong L, Ma L, Yang S, Zheng Y, Zhang J, Wu C, Zhao Y, Hou Y, Li H and Wang T: SQLE facilitates the pancreatic cancer progression via the lncRNA-TTN-AS1/miR-133b/SQLE axis. J Cell Mol Med. 26:3636–3647. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Battista MC, Guimond MO, Roberge C, Doueik AA, Fazli L, Gleave M, Sabbagh R and Gallo-Payet N: Inhibition of DHCR24/seladin-1 impairs cellular homeostasis in prostate cancer. Prostate. 70:921–933. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Fuller PJ, Alexiadis M, Jobling T and McNeilage J: Seladin-1/DHCR24 expression in normal ovary, ovarian epithelial and granulosa tumours. Clin Endocrinol (Oxf). 63:111–115. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Wu J, Guo L, Qiu X, Ren Y, Li F, Cui W and Song S: Genkwadaphnin inhibits growth and invasion in hepatocellular carcinoma by blocking DHCR24-mediated cholesterol biosynthesis and lipid rafts formation. Br J Cancer. 123:1673–1685. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Du ZP, Wu BL, Xie JJ, Lin XH, Qiu XY, Zhan XF, Wang SH, Shen JH, Li EM and Xu LY: Network analyses of gene expression following fascin knockdown in esophageal squamous cell carcinoma cells. Asian Pac J Cancer Prev. 16:5445–5451. 2015. View Article : Google Scholar : PubMed/NCBI |