Promising predictive molecular biomarkers for cervical cancer (Review)
- Authors:
- Published online on: April 4, 2024 https://doi.org/10.3892/ijmm.2024.5374
- Article Number: 50
-
Copyright: © Lizano et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
Cervical cancer (CC) is the fourth leading cause of death from cancer in women worldwide, with an estimated 604,000 new cases and 342,000 mortalities in 2020 (1,2); ~483,000 new cases and 274,000 mortalities have occurred in low- and middle-income countries (3). Vaccination and screening programs have notably reduced the incidence of CC worldwide by >80%, but this type of cancer still constitutes a serious public health problem (4). CC is considered a sexually transmitted disease because the main etiological factor associated with its development is a persistent infection with high-risk human papillomavirus (HR-HPV) (5). More than 200 genotypes have been described to date, of which ~40 genotypes can infect epithelial cells of the anogenital region (5). Of those 40, at least 13 genotypes have been defined as carcinogenic, with HPV16 being the most prevalent genotype in both squamous cell carcinoma (SCC; 59.3%) and adenocarcinoma (36.3%) (6,7).
Currently, the staging of CC is based on the criteria of the Federation International of Gynecology and Obstetrics (FIGO) and is mainly based on histological type, tumor size, infiltration of paravaginal structures and lymph node involvement (8). The accurate identification of the clinical and histopathological features of tumors is a determinant for selecting the proper therapeutic strategy (9). The treatment for early-stage disease (stages IA-IIA) includes several surgery options or a combination of pelvic radiotherapy (RT) with brachytherapy (BT) and chemotherapy, whereas the combination of cisplatin-based chemotherapy and RT, including optional application of BT, is the main therapeutic strategy employed for locally advanced CC (LACC; stages IIB-IVA) (9).
The prognosis of patients largely depends on the clinical and histopathological characteristics of the tumor at diagnosis. Women with the earliest stages of CC have a favorable prognosis, with recurrence rates <20% (stages IB-IIA); however, the risk of recurrence and/or metastasis increases up to 70 and 36%, respectively, in advanced clinical staging (stages IIB-IVB). Recurrence or metastasis notably reduces the patients' 5-year overall survival (OS) from 50-70% in stage IIB up to 40 and 10% for stages III and IV, respectively (10). Therefore, the high rate of CC mortality is strongly associated with the late diagnosis of patients with advanced-stage CC. This problem is particularly evident in low-income regions where limited coverage and access to healthcare services can prevent the timely treatment of women with advanced CC (11).
Clinical staging (tumor size) has efficiently served as a marker of chemoradiotherapy (CRT) response; however, resistance constitutes a critical issue of constant development in oncologic research, particularly its role in treatment failure (12). Clinical evidence shows that the effectiveness of CRT can differ significantly among CC patients, even among those with similar histopathological characteristics, significantly increasing the probability of recurrence and metastasis (10,12,13). This heterogeneity in CRT effectiveness highlights the need to identify new biomarkers whose prognostic or predictive values allow the selection of suitable therapeutic approaches according to individual and tumor characteristics.
Therefore, the present study reviewed the current evidence that supports the role of several proteins, methylation markers and non-coding RNAs as potential predictive biomarkers for CC. The present review also highlights the utility of participant characteristics, such as clinical stage, histological classification and lymphatic spread, for the selection of therapeutic strategies in clinical trials as an approach to precision oncology.
2. Types of biomarkers
From a clinical point of view, biomarkers are molecules whose presence, level, localization, or modification (e.g., phosphorylation, methylation and glycosylation) are useful in establishing the difference between physiological and pathological processes, determining response to pharmacological treatment, or selecting therapeutic approaches according to the underlining disease. Therefore, an ideal biomarker should be specific (whose presence, absence, or altered levels are directly associated with the development of pathological conditions or diseases), noninvasive (easy to obtain from biological fluids) and consistent with differences between ethnic groups and sexes (14).
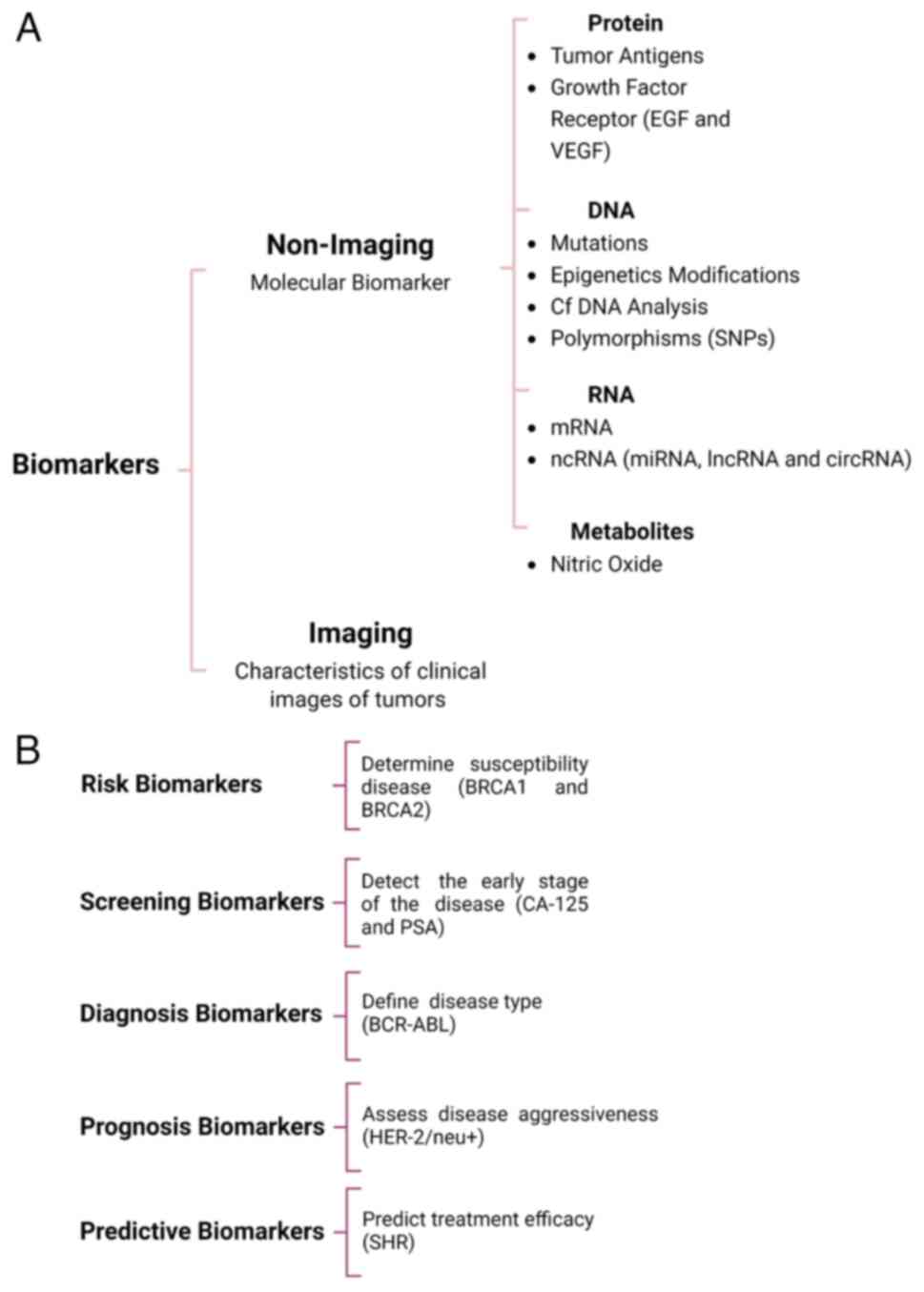
Biomarkers are classified according to several criteria, including their chemical characteristics, biological properties, application and methods employed for their detection (15). However, some biomarkers constitute components of different natures (e.g., proteins and RNAs) or participate in distinct biological processes; therefore, they can be located in various categories (16). Biomarkers can be broadly divided into imaging-based and non-imaging-based indicators (Fig. 1A). Imaging-based biomarkers focus on clinical features obtained through magnetic resonance, computed tomography and ultrasound-based techniques. The subjective interpretation and potential clinical applications of data may be limited; however, image digitalization and data analysis through algorithms (radiomics) have become essential tools for detecting tumor profiles associated with treatment failure (17,18). Non-imaging biomarkers are termed molecular biomarkers and have biophysical and biochemical properties that allow their assessment in biological samples, such as metabolites, peptides, proteins, nucleic acids and their modifications that include genetic mutations or polymorphisms, changes in methylation levels, among others (Fig. 1A) (19).
According to clinical relevance in oncology research, non-imaging biomarkers are classified into i) risk markers, assessing the susceptibility to develop a disease in an individual who currently does not present clinical manifestations; ii) screening markers that determine the probability of having the disease at an early stage; iii) diagnosis biomarkers that confirm the existence of a disease or aide in defining the clinical features of the disease; for example, biomarkers employed to determine the tumor origin and classification of cancer subtypes; iv) prognostic biomarkers that aide in determining the probability of developing a possible clinical outcome (recurrence or progression), regardless of the treatment received or in its absence (16); and v) predictive biomarkers that determine the likelihood of clinical response (favorable or unfavorable) that a patient may experience in response to specific treatment based on presence or absence of such marker. A biomarker is predictive of a favorable treatment response for patients carrying the biomarker compared with the disease course in patients without it (16) (Fig. 1B).
3. Proteins as predictive biomarkers in CC
Genetic and epigenetic changes during cervical carcinogenesis may have the potential to modify both the cell behavior and the tumor microenvironment, notably affecting tumor growth and spread (20). These genotypic changes are frequently associated with alterations to molecular expression patterns in tumors, such as non-coding RNA (small and long non-coding RNA) (21), variations in the pattern of DNA methylation (22) and modifications in protein, lipid and small metabolite (nitric oxide) levels (23), supporting their application as predictive biomarkers.
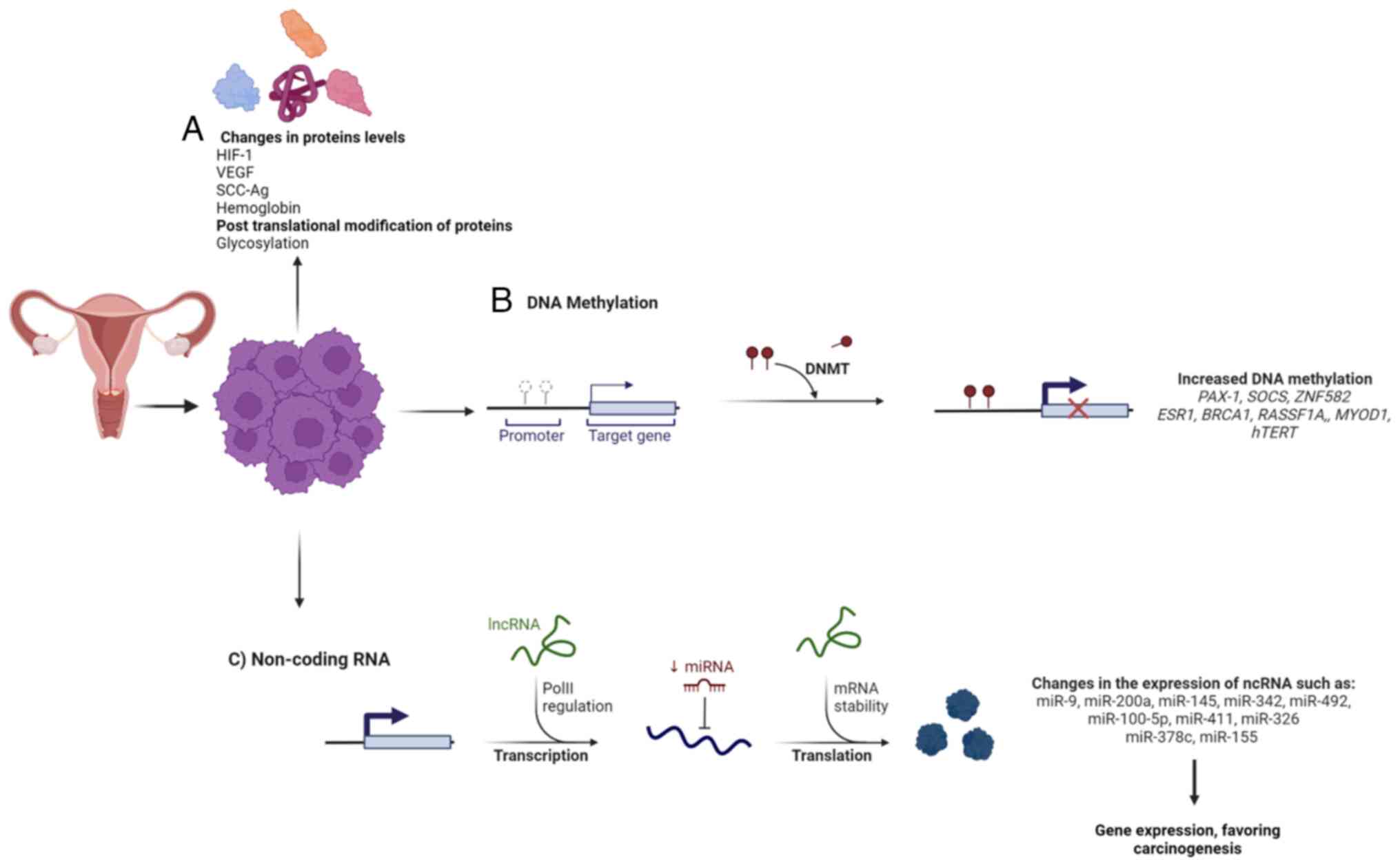
Proteins are considered the most informative biological indicator; therefore, most of the predictive biomarkers approved by the Food and Drug Administration for clinical use are proteins (Table I) (24-33). The predictive value of protein biomarkers is mainly based on their overexpression or mutations, although posttranslational modifications such as glycosylation profiles have also been identified as definitory characteristics of CC (Fig. 2A) (34).
Significant efforts have been made to determine the prognostic and predictive value of some markers in CC; however, current clinical evidence of protein markers remains controversial or insufficient for approval for clinical use (35). However, a large amount of literature has proposed candidate predictive biomarkers involved in the response to treatment, such as epidermal growth factor receptors (EGFR), apoptotic proteins (Bcl2, Bcl-2-like protein 4 and tumor protein p53), hypoxia and angiogenic factors [hypoxia-inducible factor (HIF) and vascular endothelial growth factor (VEGF)], immune checkpoints (programmed death 1 and cytotoxic T-lymphocyte antigen 4) and other proteins [cyclooxygenase-2 and squamous cell carcinoma antigen (SCC-Ag)] (36,37). This section summarizes the current evidence that highlights the strengths and weaknesses of some proteins as potential predictive biomarkers in CC.
Proteins associated with hypoxia
HIF-1 and VEGF
Tumor hypoxia is a critical factor in tumor progression and treatment resistance in solid tumors, including prostate, cervix, breast and head and neck tumors (38,39). The invasive nature of the methods employed to determine oxygenation status in tumors presents technical limitations, which has promoted the use of endogenous markers associated with tumor hypoxia, such as HIF-1, VEGF, glucose transporter 1, carbonic anhydrase IX and hemoglobin (Hgb) levels (39-41).
HIF-1 is a family of oxygen-sensitive transcriptional activators, which form dimers of α and β subunits. In hypoxic conditions, HIF-1α stabilizes and translocates into the nucleus where it binds to HIF-β to form the dimer that binds to hypoxia response elements in the promoters of genes involved in adaptation to oxygen-depletion (42-44), among which is VEGF (45). VEGF is a growth factor essential for vascular homeostasis. It belongs to a family of soluble polypeptides formed by five members (VEGF-A, B, C, D and E), of which the most studied isoform in cancer is VEGF-A because its increased levels are associated with the formation of new capillaries during carcinogenesis and metastasis (46).
The predictive value of HIF-1α, HIF-2α and VEGF is mainly associated with their increased levels in CC and their correlation with poor response to CRT (47-49). This ability to determine the sensitivity to treatment has also been evaluated in LACC patients under different treatment modalities, such as preoperative CRT (PCRT). Reduction in VEGF and HIF-1α levels were significantly associated with improved clinical responses. VEGF and HIF-1α expression rates following PCRT were 0 in patients with a complete response (CR), 70 and 63% in patients with a partial response (PR) and 100 and 85.71% in patients with stable disease, respectively. These results suggest that changes in VEGF and HIF-1α expression could predict PCRT sensitivity in LACC patients (50). This approach is particularly relevant because PCRT improves progression-free survival compared with that associated with RT (92 vs. 76.5%) (51). Moreover, PCRT provides other clinical advantages, such as the opportunity for subsequent radical surgery, the identification of the response rate and adverse effects of CRT before determining a treatment plan and the opportunity to retain ovarian function in young patients (52,53). The changes observed in VEGF protein levels are similar to the findings observed in patients with LACC before PCRT: the reduction of VEGF levels after PCRT (P<0.001) significantly correlated with CR in 82.2% of participants, with a 3-year progression-free survival (PFS) of 84.8% (Table II) (54).
Platinum-based neoadjuvant chemotherapy (NACT) before radical hysterectomy or radiotherapy has shown favorable response rates of OS and PFS in LACC patients (55). This therapeutic strategy is associated with a significant reduction in tumor size, which may improve prognosis by reducing the risk of recurrence and micrometastasis (56). However, the lack of clinical response implies the indication for postoperative radiation, which may delay the treatment options and worsen the prognosis (56). Therefore, the identification of biomarkers that can predict a clinical response to NACT could optimize therapeutic options for LACC patients. Consistent with this proposal, findings reported by Zhao et al (57) in a group of 42 LACC patients treated with NACT pre-hysterectomy showed that high levels of HIF-1α and VEGF protein significantly correlated with improved clinical responses to NACT. The reduction of HIF-1α and VEGF post-NACT was associated with a CR (Table II). These results suggest that protein levels of HIF-1α and VEGF-A may guide the selection of PCRT and NACT in patients because the reduction is associated with a higher probability of improved clinical responses. However, the predictive value of these markers seems to be limited to LACC because higher protein levels in patients with early-stage cancer correlated with a negative response to treatment with NACT (57,58).
It is important to highlight that the clinical utility depends on VEGF and HIF-1α not only on the type of sample (serum or tissue) but also on the accurate diagnostic of the methods employed for detection, which determines the establishment of cut-off points. Due to the reduced size and the limited follow-up of the study population, in addition to differences in methods employed for their detection, further studies are required to determine the sensitivity and specificity of VEGF and HIF-1α for predicting tumor recurrence and treatment response in patients with LACC, particularly employing methods whose principle of detection allows a comparative analysis.
Hgb
Low oxygenation in solid tumors increases the risk of local invasion, metastasis and treatment failure (59,60). Hgb level is considered a determining factor for clinical response during treatment owing to Hgb's role in oxygen transportation. Higher oxygenation levels are reached when Hgb ranges between 12-14 g/dl, whereas lower levels of Hgb are associated with tumor hypoxia and anemia in the patient (40,61). Most clinical studies on CC support the role of Hgb levels as a prognostic biomarker (61,62); however, in some studies pretreatment Hgb levels were not significantly correlated with clinical response in patients treated with concurrent chemoradiotherapy (CCRT), so the predictive value of Hgb remains controversial (40,63).
According to Gennigens et al (63) the follow-up of patients with LACC, during treatment with cisplatin-based chemoradiotherapy, revealed a significant decrease in Hgb levels (<12 g/dl) compared with the median Hb at diagnosis (13.1 g/dl). However, no significant association was observed with OS and recurrence-free survival (RFS). This association is consistent with a cohort of LACC patients treated with different therapeutic strategies, which demonstrates that both RT and CRT can be associated with a significant reduction in Hgb levels during treatment (40). However, only Hgb levels <10 g/dl were negatively associated with a reduction in disease-specific survival. Moreover, transfusions showed no significant effect in improving the effectiveness of the treatment with CCRT but worsened outcomes in patients treated with RT alone (40). These findings suggest that a pretreatment Hgb level <11 g/dl may be associated with a lack of response in LACC patients treated exclusively with RT, which is in line with results reported by Moreno-Acosta et al (64). Pretreatment Hgb level >12.7 g/dl was significantly associated with CR to RT in LACC patients (P<0.001), which significantly increased the OS and DFS of the cohort.
These results highlight the usefulness of considering anemia to predict the risk of response failure in patients with LACC receiving RT. However, the retrospective nature of most of these studies can cause potential biases. Although all patients met several strict inclusion and exclusion criteria to constitute a homogeneous group of patients, further prospective research is necessary to confirm the usefulness of this new biomarker.
Other hematologic biomarkers may also be needed to determine which patients are at a particular risk of treatment failure, requiring closer monitoring and intensified systemic treatments. The changes in polymorphonuclear neutrophils and white blood cells between the first and last CT cycles may be interesting new biomarkers, as they correlate with RFS, OS and distant recurrence (65). It could be combined with other well-known pre-treatment predictive markers (tumor size and nodal disease) to anticipate adjuvant systemic treatments (CT or new drugs, such as immunotherapy) if ongoing trials prospectively confirm its positive impact.
SCC-Ag
The serum SCC-Ag is a glycoprotein derived from tumor squamous cells and expressed through two isoforms: SCC-Ag 1 and SCC-Ag 2 (66). The expression of SCC-Ag 1 is mainly associated with radiation resistance. Therefore, elevated SCC-Ag levels may predict treatment failure in SCC patients (67,68). The literature highlighting the clinical findings supporting this hypothesis is described in the following section.
Hysterectomy (combined with adjuvant RT) or CCRT are equally effective treatments for patients with early stages of CC (IB1-IIA) (9). However, surgery is only offered for patients with a low probability of undergoing adjuvant CCRT or RT to avoid using two different treatments. Therefore, an adequate preoperative assessment of the risk factors is critical to determine the therapeutic strategy and can predict the response treatment (69). Lymph node metastases (LNM) are routes of CC dissemination. LNM is a poor prognostic feature, even in patients with early-stage disease (70). The evaluation of preoperative LNM is based on medical imaging, which has limitations. Therefore, it is imperative to find new markers that, together with the data obtained by imaging, allow the prediction of the presence of LNM for selecting treatments for patients with early-stage CC. Xu et al (69) determined that preoperative SCC-Ag levels >2.35 ng/ml could be used as a predictor of LNM. Moreover, combination with computed tomography generated reliable data for predicting LNM, with a sensitivity of 82.9%, although the specificity decreased according to the severity of CC owing to its increased levels consistently observed in advanced stages (69). These results support the potential of SCC-Ag as a predictive marker for the use of definitive RT, with reliable clinical results in patients with early-stage SCC (Table II).
Regarding cases of LACC treated with definitive CCRT, Choi et al (68) reported in a cohort of 304 SCC patients that pretreatment SCC-Ag levels ≥4 ng/ml significantly correlated with higher local, regional and distant metastasis rates. Similar results were reported by Kang et al (71) who showed that SCC-Ag levels (>2 ng/ml) before CCRT were predictive of the development of distant recurrence within 5 years. These results show that increased SCC-Ag levels before treatment could be useful in predicting treatment failure and selecting adjuvant therapies, such as neoadjuvant chemotherapy, consolidation chemotherapy and extended-field radiotherapy (EFRT), in patients treated with definitive CCRT (72) (Table II).
The lymphatic spread in CC follows a predictable and orderly pattern from the lower to the upper regions of the pelvis. Para-aortic lymph nodes play an important role in the metastases of CC (73). According to the Gynecologic Oncology Group, 25% of patients with LACC have para-aortic node metastasis (PAN), which is associated with poor survival rates (74). In these cases, EFRT plus CCRT (EF-CCRT) is recommended postoperatively to provide suitable loco-regional disease control (75,76). However, distant metastases are the leading cause of failure of treatment. Clinical trials have determined that postoperative patients with PAN who underwent adjuvant chemotherapy with EF-CCRT had a lower incidence of distant metastases than patients who received EF-CCRT alone (77,78). The critical issue is identifying patients who have undergone postoperative EF-CCRT and can benefit from consolidation chemotherapy. In this condition, SCC-Ag levels could be an essential biomarker since previous studies have shown that 70-86% of patients with metastasis had elevated SCC-Ag levels (≥4 ng/ml) (67,68,79).
Zhang et al (80) demonstrated the predictive value of pretreatment serum SCC-Ag levels in a cohort of 113 patients who underwent postoperative EF-CCRT. Serum SCC-Ag levels were analyzed in 63 patients who received EF-CCRT and consolidation chemotherapy and another 50 patients who received EF-CCRT alone. The results showed that patients with low pretreatment levels of SCC-Ag did not experience a change in clinical outcome when they received consolidation chemotherapy, compared with the outcomes of those who did not. However, in patients with pretreatment serum SCC-Ag >6.5 ng/ml, adjuvant chemotherapy demonstrated significantly decreased systemic recurrences. Similar results were observed in previous studies (68,80).
Changes in SSC-Ag levels during treatment are hypothesized to have the potential to aid in the follow-up of patients, given that the regression of tumors after CCRT may require >3 months (79) and the difficulty in identifying whether a patient has achieved or will achieve a CR via gynecologic examination. According to Kawaguchi et al (81), the changes and the establishment of cut-off levels could be useful in predicting the risk of recurrence or metastasis according to the therapeutic approach (RT or CCRT). Posttreatment SCC-Ag levels >1.15 ng/ml or 1.20 ng/ml were associated with decreased 3-year OS in LACC patients treated with CCRT (84%) or RT (95%), respectively. However, its application depends on the therapeutic approach as well as the clinical staging of tumors, because post-treatment SCC-Ag levels >1.0 ng/ml can be associated with a higher risk of recurrence and an incomplete response to RT in patients with stage IB to IIIB disease (82).
Serum biomarkers such as SCC-Ag are essential in cancer treatment because serum is an ideal type of clinical sample due to its easy accessibility, low cost and non-invasive nature. Furthermore, the level of SCC-Ag is increased in 88% of SCC patients compared with healthy women (83). In addition to CC, SCC-Ag has been proposed as a clinical tool to implement personalized treatment policies for cell carcinomas of organs such as the tongue, esophagus, tonsils and lungs (non-small cell lung cancer) (84,85). However, the clinical relevance of SCC-Ag in cancer treatment and prognosis remains controversial because some evidence (such as those analyzed in the present study) supports the fundamental role of SCC-Ag as a predictive biomarker in CC. However, on the contrary, other authors demonstrated that the evaluation of SCC-Ag during follow-up did not improve the response to treatment or the early detection of the recurrence (86). SCC-Ag must be validated as a predictive biomarker to be incorporated into clinical practice, which implies the homogeneous use of highly sensitive technology and defined criteria for patient eligibility. SCC-Ag has not been documented in the current guidelines for routine clinical use in patients with cancer. To this end, it is necessary to conduct prospective and large-sample studies that allow the establishment of reliable cutoff levels of SCC-Ag that will help in clinical decision-making. It is advisable to evaluate combinations with other markers, including tumor characteristics, such as size, parametrial involvement and lymph node enlargement.
4. DNA methylation markers
Genomic instability is a hallmark feature of cancer cells, correlating with the increased risk of acquired mutations. Therefore, mutations of a single nucleotide (single nucleotide variants) are popular prognostic and predictive biomarkers in cancer (87). However, alterations in DNA methylation status are also considered determinants of genomic instability and multiple efforts have been directed to determine their possible role as biomarkers either in diagnosis or clinical response to cancer treatment (Fig. 2B) (88,89). DNA methylation refers to the covalent addition of methyl groups to carbon 5 of cytosine rings, specifically in the CpG island promoters (promoter hypermethylation) of several genes (90). Changes in levels and patterns of DNA methylation are frequently associated with the aberrant expression of genes that actively contribute to carcinogenesis by mainly regulating cellular growth and differentiation (91). Alterations in DNA methylation play a determining role in maintaining a malignant phenotype and their prognostic and predictive performance has been evaluated in different cancers, such as ovarian, gastric, breast, esophageal, glioblastoma, colon, melanoma and lung cancers (88,92,93). Currently, methylation-based biomarkers approved by the United States Food and Drug Administration (FDA) include the qualitative evaluation of Septin 9 gene methylation for the diagnosis of colorectal cancer in plasma samples (Epi ProColon), the combination of N-Myc Downstream-Regulated Gene 4 and Bone Morphogenetic Protein 3 (ColoGuard) for the diagnosis of colorectal cancer in stool samples (88) and O6-methylguanine-DNA methyltransferase methylation as a prognostic and predictive marker in glioblastoma multiforme (94).
The contribution of DNA methylation to the alteration of genes associated with the development and progression of CC suggests that changes in DNA methylation could be employed for selecting therapeutic approaches with lower risks of recurrence, progression, or metastasis (95-97). However, few studies have explored the usefulness of changes in DNA methylation status for predicting the clinical response of CC treatments. Clinical studies that evaluated DNA methylation as a biomarker for CC diagnosis (98,99) show that methylation analyses of two or more genes and cutoff limits could be main factors that modify their predictive value.
The evaluation of changes in DNA methylation through semi-quantitative assays has suggested that combined hypermethylation of Myogenic Differentiation1 (MYOD1), Estrogen Receptor 1 (ESR1) and human telomerase reverse transcriptase (hTERT) could be associated with CR in patients with LACC (stage IIB/III) receiving CRT (100). However, according to Contreras-Romero et al (101), differential methylation of the Bromodomain Containing 9 and the Cytosolic Thiouridylase Subunit 1 was significantly associated with improved OS and PFS and in patients with LACC (stage II/III-V), whereas hypomethylation of Dedicator of Cytokinesis 8 correlated with shorter OS and resistance to CCRT. On the other hand, Guerrero-Setas et al (102) suggested that hypermethylation of Ras association domain family member 2 (RASSF2) could be sufficient to determine the risk of recurrence by reporting that RASSF2 hypermethylation before treatment was significantly associated with high likelihood of recurrence in patients with LACC (stage II/III-V) treated with CCRT (102). These results indicate that the predictive value of the biomarker might be significantly modified by the heterogeneity of samples and the sensitivity of the method employed for the determination of DNA methylation.
Methylation of paired-box transcription factor 1 (PAX1), a protein that plays a critical role during embryogenesis, has shown consistent results throughout multiple studies (103). PAX1 acts as a tumor suppressor in different cancers (cervical, ovarian, colorectal carcinoma, parathyroid and oral SCC) (104). Methylation levels of PAX1 increased according to the severity of cervical lesions; therefore, its predictive value has been mainly evaluated in patients with LACC receiving CCRT. Experimental assay and analysis of genome-wide methylation through algorithms showed that PAX1 expression can be regulated by hypermethylation of its promoter in CC cell lines; furthermore, increased PAX1 methylation is significantly associated with shorter OS and PFS in patients with LACC (105). However, methylation level changes in PAX1, that were evaluated using relative quantification (ratio of PAX1/COL2A methylation), were associated with an improved response in patients treated with RT. The reduction in methylation levels of PAX1 in stage IIB/IIIB tumors during the RT phase was significantly correlated with smaller tumor size, which was observed in patients with CR (106). An analysis of cutoff values (delta CP ≤9) showed that differences in PAX1 methylation, observed during the RT phase, could predict a failed response to treatment with high sensitivity (72%) and specificity (88%). These findings highlight the importance of cut-off values for determining the predictive ability of the marker, supporting the proposal that changes in the methylation status of the PAX1 gene could be used as a predictive biomarker of early response in CC patients treated with radiation. However, further prospective studies are required to determine the reliability of PAX1 methylation for predicting the response to RT. Furthermore, assessment of PAX1 methylation using absolute quantification methods may be useful in determining whether cut-off values can be applied to all populations or in identifying technical factors modifying the accuracy and precision of the test.
Some studies have also proposed that methylation of zinc finger protein 582 (ZNF582) could be employed to evaluate the clinical response in patients with LACC treated with CCRT (107,108). ZNF582 is a member of the family of zinc finger proteins that function as transcription factors and is involved in several cellular processes such as DNA damage response, proliferation and cell cycle regulation factors. Decreased expression of ZNF582 is frequently associated with promoter hypermethylation in several types of cancer such as esophageal SCC, anal cancer, colorectal cancer and CC (108-110). Moreover, methylation levels of ZNF582 increase according to the severity of cervical alterations (110). These findings support the active participation of ZNF582 methylation in cervical carcinogenesis, suggesting that the determination of its methylation status could be applied to predict the response to therapeutic strategies employed in invasive CC. According to Wu et al (111), the predictive capability of ZNF582 methylation in RT was evaluated in patients with LACC treated with neoadjuvant CRT (NCRT) where 78% of patients had tumor FIGO staging <IIB and 88.3% had glandular types. Methylation levels of ZNF582 lower than the cut-off values (ratio of ZNF582/COL2A methylation, delta CP ≤13) showed significantly improved clinical outcomes with NCRT. Methylation levels lower than the cutoff values induced following NCRT were significantly correlated with an increased risk of short-term recurrence compared with patients with ZNF582 hypermethylation. Conversely, hypermethylation of ZNF582 after NCRT was associated with a higher rate of 5-year disease-free survival in comparison to that in patients with unmethylated ZNF582 (84.5 vs. 72.4%; P=0.04). Therefore, these results suggest that ZNF582 methylation could be considered a predictive biomarker in cervical adenocarcinoma. However, the predictive value of ZNF582 methylation was found to be closely associated with the methods (standard curve or relative quantification) and cutoff values employed to determine the hypermethylation status, which determines the sensitivity and specificity of the test (108,111). Therefore, further studies with homogeneous criteria, such as absolute quantification, are required to clarify the role of methylation levels in responses to radiation.
Evaluation of data from public repositories has also contributed to the determination of the predictive behavior of DNA methylation (112). This approach allows the application of different algorithms for data analysis. DNA methylation is usually associated with the inhibition of gene expression; therefore, the correlation between methylation and expression values provided by the algorithms supports the biological effects of DNA methylation on gene expression. The predictive model has shown that differential methylation of Zinc finger FYVE domain-containing protein 21, Cytochrome B-245 Chaperone 1, protein disulfide isomerase family A member 6, Cullin 7, T-cell surface glycoprotein CD3 epsilon chain, T cell immunoreceptor with Ig and ITIM domains, M-phase phosphoprotein 10 and LOC100132215 could predict a higher risk of recurrence in patients with LACC (113). These findings show that the use of algorithms for methylation status associated with dysregulated genes in CC may be a useful tool to identify whether changes in methylated regions could be associated with sensitivity to treatment; however, studies with a larger number of samples are needed to strengthen these results.
5. Non-coding RNAs as predictive biomarkers in CC
Understanding the genetic composition of cells and their regulation has progressed rapidly since the development of human genome sequencing. Only ~1% of the human genome is coding DNA; however, ~90% of the genome is transcribed, which implies that a large proportion of these non-coding transcripts have important regulatory purposes in different cellular contexts (114). Much has been learned about the several types of non-coding RNA (ncRNAs); however, their action mechanisms and cellular targets are not completely known.
ncRNAs are traditionally classified into two types according to the size of the transcript: Small non-coding RNAs (20-200 nt) and long non-coding RNAs (>200 nt). Nevertheless, a wide, heterogeneous variety of ncRNAs have complex regulatory and structural functions. Several classes of ncRNAs have been identified, including transfer RNAs (tRNAs) and tRNA-derived small RNAs (tDRs or tRFs) (115); ribosomal RNAs (rRNAs) (116), microRNAs (miRNAs) (117); small interfering RNAs (siRNAs) (118); small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) (119), circular RNAs (circRNAs) (120); long ncRNAs (lncRNAs) and transposable elements (121,122); piwi-interacting RNAs (piRNAs) (123); Y RNA (124); and vault particle-associated RNAs (vtRNAs) (125).
These ncRNA groups can perform different roles in biological functions or underlying pathological conditions, acting as tumor suppressor genes or oncogenes that participate in or regulate malignant tumor development. The utility of ncRNAs for diagnosis and progression and as therapeutic targets and predictive biomarkers has been widely explored owing to their high degree of conservation, specific expression patterns and stability (126-128). miRNAs, circRNAs, lncRNAs, siRNAs and piRNAs have been shown to play crucial roles as regulators of the response to chemotherapy, immunotherapy and molecular-targeted therapy in different types of cancer (117,118,126,129,130).
Several studies on CC have identified the role of ncRNAs as prognostic biomarkers; however, limited information is available regarding their use in predicting treatment response. Therefore, the present study sought to include information on ncRNAs as biomarkers with predictive value in CC, as determined by studies carried out not only in cell lines, but also in patient samples.
miRNAs
Among the small non-coding RNAs, the best characterized are siRNAs and miRNAs. The two participate in post-translation regulation mechanisms associated with gene silencing, either by mRNA degradation or interfering with the mRNA translation process (Fig. 2C) (108,117). Furthermore, miRNAs, when secreted in exosomes, can function as ligands of Toll-like receptors (TLRs), which activate TLR signal transduction pathways and induce the secretion of pro-metastatic inflammatory cytokines (131). miRNAs have also been shown to function in the nucleus by regulating gene transcription or targeting mRNA splicing, as well as preventing RNA export (132). Growing evidence indicates the aberrant expression of miRNA is strongly associated with the development and maintenance of malignant phenotypes of CC cells. Moreover, miRNAs have been employed as molecular indicators of recurrence, progression and metastasis in patients with ICC (133). Table III shows miRNAs involved in CC treatment response.
Due to the great diversity of miRNAs, profiles with different combinations of miRNAs have been identified to help predict therapeutic response in CC. Some studies evaluated the predictive value of mRNA tumor profiles obtained from CC (stage I/II/III) in the response to CRT. According to Hu et al (134), the generation of a predictive model (algorithm) based on the correlation between miRNAs and OS allows the identification of miRNAs directly associated with a lower risk of therapy resistance (TR). This model showed that changes in miR-200a and miR-9 before radiation were significantly associated with a higher RFS rate (P=0.036). Moreover, the predictive value of mi-R200a has also been proposed by Pedroza-Torres et al (135) in their cohort of patients with LACC (stage II/III) treated with CCRT (pelvic radiation + BT). The reduced pretreatment levels of miR-200a observed in non-responding patients negatively correlated with the risk of TR [underexpressed 5.82-fold in non-responders compared with responders (P<0.001)].
The predictive value of certain miRNAs has also been explored using indicators that have shown prognostic utility in CC. Regarding miR-145, changes observed during CC development and progression have been associated with poor prognosis and shorter OS. The reduction of miRNA-145 levels observed in CC before CRT is more pronounced in patients with poorly differentiated tumors (P=0.027), LNM (P=0.009) and advanced tumor stage (P=0.002) (136,137). These findings have supported the potential application of miR-145 in predicting the clinical outcome of radiotherapy in patients with LACC. According to Wei et al (138), the plasma levels of miR-145 negatively correlated with the severity of CC. Moreover, pretreatment plasma miR-145 levels of patients with CR were significantly higher compared with those of non-responding patients (P=0.005). Accordingly, plasma miR-145 levels could be employed to determine the radiosensitivity in patients with LACC, with high specificity (84.6%). The association of miR-145 with chemotherapy resistance has not been explored in clinical studies; nevertheless, experimental assays showed that a reduction in miR-145 levels is associated with increased expression of E6 oncogene of HPV18 induced by cortisol, reducing mitomycin-induced apoptosis in the HeLa cell line. Conversely, miR-145 overexpression reverses glucocorticoid-induced chemoresistance, which suggests that changes in the levels of miR-145 might be associated with chemotherapy resistance (139). However clinical studies are required to determine its predictive value of miR-145 in patients treated with chemotherapy.
Other miRNAs showing promising predictive values include miR-492, miR-411, miR-100-5p and miR-326. Increased expression of miR-492 in patients with LACC was observed in patients responding to CCRT (140). Notably, increased miR-492 levels in patients with positive LNM show a high predictive value, with a 75.0% specificity and 95.24% sensitivity (140). These findings were further studied with in vitro assays, which showed overexpression of miR-492 turned SiHa cells radiosensitive, increasing the fraction of apoptotic cells (140). Clinical evidence supporting the role of miR-100-5p as a predictive biomarker in LACC patients, show increased expression of miR-100-5p in pretreated patients who have had a complete response to CCRT (141).
Regarding miR-411, its downregulation is significantly associated with a low probability of response to RT in CC patients (stage I/II/III). Higher expression of miRNA-411 is observed in responding patients (CR and PR) compared with that observed in non-responders (PD and SD) to RT (P<0.05). This response is consistently observed in tumor and blood samples, which highlights its utility as a predictive non-invasive marker (142). According to Zou et al (133), the change in expression of miR-326 levels detected in the serum of patients with LACC is significantly associated with an improved response to NACT. miR-326 levels in responding groups show a significant increment after treatment, whereas those of the non-responding group remained unchanged (P<0.023; sensitivity: 88.89%; specificity: 50%).
Analysis of the data set (The Cancer Genome Atlas repositories) generated by methods of high-throughput sequencing, such as RNAseq, has provided valuable information about the tumor profile of miRNAs associated with response to platinum treatment. The expression of hsa-miR-342 (P=9.68×10-5), hsa-miR-378c (P=1.29×10-4) and hsa-miR-155 (P=2.01×10-4) is significantly associated with response to platinum-based (cisplatin or carboplatin) treatment in a group of 94 SCC samples; however, the reduced number of cases of CC employed for the predictive model (logistic regression) did not allow the assessment of their predictive value (137). These high-through sequencing methods have also been employed to determine if the resistance to CRT could be associated with tumor profiles of miRNAs originating from CC stem cells (CCSC). It has been proposed that the heterogeneous composition of malignant tumors is a characteristic associated with the plasticity and aggressiveness of CCSC, which increase the risk of TR. In this regard, Zuccherato et al (143) analyzed the transcriptome of a stem cell-enriched population from CC samples from cases of responders and non-responders (NR) to chemoradiotherapy. Notably, among diverse genes and ncRNAs, they found four miRNAs that were overexpressed in NR (miR-4278, miR-4422, miR-4779 and miR-1268B), with 16 putative targets with a significant negative correlation identified by binding prediction. With this analysis, the authors define a gene/miRNA expression profile associated with NR patients and a worse prognosis.
Due to changes in plasma levels of certain miRNAs correlated with alterations observed in the primary tumor, the evaluation of their levels in blood has been considered a non-invasive method for the follow-up of patients during treatment (136,138,142). Therefore, their use is being analyzed as biomarkers during progression and as predictors of treatment response and their participation in the development of cancer has been highlighted (144,145). However, more studies with rigorous analyses should be conducted to determine the strongest candidates for use as predictive biomarkers.
Moreover, several signaling pathways have been implicated in radioresistance in cervical cancer, such as the Hedgehog pathway (146); Wnt/b-catenin pathway (147) and MAPK pathway (148), among others; which may be the target of the different aforementioned miRNAs. Several mechanisms have been proposed by which miRNAs contribute to chemotherapy resistance in CC, particularly to cisplatin, such as apoptosis inhibition, regulation of mismatch repair system, rise in epithelial-mesenchymal transition characteristics, increase in the number of drug-resistant cancer stem cells following chemotherapy and the reduction of oxidant levels (149). For example, miR-92b suppresses the LDLRPTEN signaling pathway, leading to activation of the AKT pathway, which inhibits apoptosis (150); miR-7-5p negatively regulates Poly (ADP-ribose) polymerase 1 (PARP-1), which is an important regulator of the mismatch repair mechanisms (151).
Understanding the mechanisms by which these miRNAs, through their target genes, are associated with treatment response is an aspect that deserves further study. This would allow the identification of molecular targets that regulate key signaling pathways in tumor progression and recurrence, against which targeted therapies could be sought.
Long non-coding RNAs (lncRNAs)
lncRNAs are longer than 200 nucleotides and are not translated into proteins (152). lncRNAs can interact with RNA, DNA and proteins according to the different biological processes where they are expressed (153,154). Based on their gene location, lncRNAs can be classified into five categories: Long intergenic ncRNAs, long intronic ncRNAs, sense lncRNAs, antisense lncRNAs and bidirectional lncRNAs. In line with their regulatory effect on DNA sequences, lncRNAs can be classified as cisor trans-acting lncRNAs (155,156).
lncRNAs involved in tumor progression and invasion that have been considered as markers of therapeutic resistance include HOX transcript antisense RNA (HOTAIR), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), H19, colon cancer-associated transcript 2, Sprouty4-Intron 1, growth arrest-specific 5 (GAS5), maternally expressed 3 and prostate cancer-associated transcript 1 (PCAT1) (157,158). However, some of them have also been considered diagnostic and prognosis biomarkers and therapeutic targets for CC treatment owing to their roles in CC development, progression and metastasis (21,159,160).
Increasing evidence has shown that increased expression of HOTAIR may be useful to predict response to RT in CC. Results obtained from biological models suggest that overexpression of HOTAIR increases the risk of radioresistance in CC by upregulating of HIF-1α (161). According to Zhou et al (158), the negative correlation between HOTAIR and miR-214-3p could be associated with the alteration of genes involved in the epithelial-mesenchymal transition (Wnt/β-catenin pathway), which may also contribute to the acquisition of radioresistance in CC cells. Moreover, the majority of clinical evidence indicates that the increased levels of HOTAIR observed in CC patients are associated with tumor size and increased probability of LNM, which significantly reduces the OS of patients treated with CCRT (162). These findings highlight the value of the HOTAIR expression as a prognostic marker; however, further prospective studies are required to determine its predictive performance in patients with LACC treated with different therapeutic strategies.
A number of experimental studies on the biological mechanisms of lncRNAs involved in the response of CC to treatment have been published; however, the present study mainly focused on the revision of studies showing experimental findings and clinical evidence supporting the predictive performance of the markers. In this regard, Lu et al (163) show that MALAT1 levels before treatment are significantly higher in patients with radioresistance compared with those of responding patients. The study also evaluated the regulatory effect of MALAT1 on miR-145 (competing endogenous RNA mechanism) and a negative correlation was observed in the tumor samples (R2=0.52). These findings were obtained in a cohort of patients with LACC (IIB/IIIA) treated with CCRT (pelvic radiation + BT). Moreover, experimental assays in HPV-positive cancer cell lines showed that inhibition of MALAT1 and overexpression of miR-145 significantly reduces cell proliferation and increases sensitivity to radiation-induced apoptosis (163). Together, these results highlight the synergism of MALAT1 and miR-145 in the development of radioresistance during CC treatment.
A similar approach was employed to evaluate whether the relationship between GAS5 and miR-106b could be associated with an improved response of CC cells to radiation. According to Gao et al (164), pretreatment expression levels of GAS5 and miR-106b in patients with LACC (IIB/IVB) can predict the response to radiotherapy. Although the ratio between both markers is not analyzed, lower GAS5 and higher miR-106b levels are observed in patients not responding to RT compared with those observed in the responding group. These results are consistent with GAS5 and miR-106 levels observed in the CC cell line (SiHa) showing radioresistance. Therefore, the response to radiation was determined by pretreatment GAS5 levels, which directly and indirectly regulated the expression of miR-106b and immediate early response 3 (IER3), respectively. On the other hand, the effectiveness of GAS5 for predicting the response to chemotherapy has also been evaluated in a cohort of CC patients (stages I-IV). Lower GAS5 levels significantly correlate with resistance to cisplatin treatment and a significantly shorter OS (165); however, the population analyzed was small. It is necessary to increase the study population to confirm whether the expression levels of GAS5 could be useful in predicting the response CCRT.
Although the effectiveness of PCAT1 for determining the response to CC treatment has not been explored in clinical studies, the aberrant expression detected in LACC indicates its possible role as a predictive marker. According to Ge et al (166), higher PCAT1 levels are detected in advanced stages of CC (III-IV) compared with those observed in normal tissue, which is consistent with in vitro evaluation of PCAT1 in CC cell lines. The relevance of PCAT1 in the response to RT is complemented by the evaluation of miR-128 and GOLM1 expression in tumor xenograft assay treated with radiation (X-rays). The inhibition of PCAT1 in HeLa xenotransplanted cells result in a significant reduction in tumor volume following radiation. Therefore, the authors propose that the sensitization of CC cells to radiation may be indicated by the axis of action of PCAT1-miR-128-GOLM1 (166).
Circular RNAs (CircRNAs)
CircRNAs are RNA molecules characterized by a continuous loop structure and are covalently closed at their free 3' and 5' terminal ends, which gives them their characteristic of being highly conserved and stable within the cell. They are derived from a precursor messenger RNA (pre-mRNA) either by back-splicing or canonical splicing (120,167,168). CircRNAs lack an internal ribosome entry site and cannot be translated (169,170); however, they regulate a great variety of cellular mechanisms such as gene expression, protein translation and cellular signaling, among others, and aberrant expression is frequently associated with the development of some human diseases, particularly types of cancer (171-174). Notably, an analysis of the expression landscape of circRNAs in human cancer cell lines has revealed distinct tissue-specific expression profiles (175), suggesting that circRNAs can be used as diagnostic, prognostic, or predictive markers for different types of cancer. The covalently closed circular structure of circRNAs renders them resistant to exoribonucleases (176), which is advantageous when considering their usefulness as possible prognostic or predictive biomarkers.
CircRNAs exert their regulatory function through three main mechanisms: As sponges for miRNAs, forming a complex with circRNA-binding proteins, or serving as a template for protein synthesis (177,178). In addition, circRNAs actively participate in the acquisition and development of the metastatic potential of cancer cells, modifying the response to therapeutic options (179,180). Therefore, circRNAs have been implicated in tumor progression and treatment response in several types of cancer such as breast cancer, lung cancer, hepatocellular carcinoma and gastric cancer (181-184). Several studies have shown that some circRNAs are efficiently translated into peptides (185,186) and the functional aspects of the translated peptides have also been studied (171).
Some studies have proposed that the aberrant expression of circRNAs is strongly associated with CC progression (187-189). The contribution of circRNAs to the acquisition and maintenance of malignant phenotypes during cervical carcinogenesis support their usefulness as diagnostic biomarkers; for example, the combined analysis of hsa_circ_0101996 and hsa_circ_0101119 in the peripheral whole blood determines the probability of LACC, with sensitivity (94.3%) and specificity (87.3%) (190). Other studies identify the contribution of circYPEL2 and hsa_circ_0065898 in mechanisms related to progression, recurrence and metastasis during treatment, highlighting their utility for diagnosis, prognosis and as molecular targets in SCC (191,192). However, clinical information regarding the predictive performance of circRNAs in CC treatment is limited.
Alterations in the expression of circEPSTI1 have been employed to elucidate the mechanisms associated with resistance to chemotherapy in CC patients. The comparative expression of cervical tissues show that circEPSTI1 levels are significantly higher in CC tissue compared with their levels in the adjacent normal tissue. Although clinical data of patients were not available for comparison with response to chemotherapy, in vitro assays show that the inhibition of circEPSTI1 in cell lines increases their sensitivity to cisplatin. This response is partly explained by the sponging effect of circEPSTI1 on miR-370-3p and the indirect regulation of mutS homolog 2 (MSH2), a gene involved in DNA damage repair (193). These findings suggest that increased circEPSTI1 in CC patients may be associated with an increased likelihood of chemotherapy failure but also highlight its usefulness as a therapeutic target to overcome resistance.
Due to the clinical relevance of paclitaxel (PTX) in CC treatment, either as NCRT or in combination therapy with cisplatin, some studies have evaluated the predictive performance of circRNAs in the PTX response. Regarding circZFR (hsa-circ 0072088), resistance to PTX in CC patients significantly correlates with higher levels of circZFR compared with the non-responding group, which negatively correlates with the OS of patients. According to experimental results, the induction of PTX resistance in CC cell lines increases circZFR levels, which is associated with the modulation of the miR-944/IL-10 axis. Some studies have proposed other responding mechanisms based on the increased expression of circRNAs in patients with PTX resistance, e.g., the regulation of circCEP128 on miR-432-5p and circMYBL2 on miR-665 (194,195). Conversely, the regulation of hsa_circ_0009035 on miR-889-3p may be associated with sensitivity to RT (196). The increased expression of hsa_circ_0009035 in CC patients not responding to RT supports its possible use as a target therapeutic and predictive biomarker. However, further prospective studies are required to clearly define the clinical and histopathological characteristics of individuals that provide a greater effectiveness of predictive markers.
Current information shows that ncRNAs, particularly miRNAs, lncRNAs and circRNAs, are being positioned as biomarkers with predictive potential for the treatment of CC. However, further studies are required to determine the advantages of these biomolecules over existing markers in predicting the clinical outcomes of patients undergoing current treatments for cervical cancer.
6. Future applications of predictive biomarkers
Currently, the predominant model in the treatment of patients with CC includes surgery, chemotherapy and radiotherapy. However, the efficacy rate of predefined therapeutic approaches varies depending on the histopathological characteristics of patients and poor clinical response is often observed (9). This situation suggests that the massive application of these therapeutic approaches not only implies the exposure of treatments that may not generate the expected clinical benefit but could also reduce the probability of a successful response to a personalized treatment owing to the toxicity of the multiple treatment schemes employed. Key precision oncology relies on an integrated insight into the genomic, transcriptomic and proteomic background of the patients to establish biomarkers with sufficient sensitivity and specificity to guide clinical decision-making.
Although numerous studies have supported the usefulness of nucleic acids and some proteins as predictive biomarkers, most of them have yet to exhibit the necessary foundation for their clinical application. Even studies investigating the predictive utility of the combined markers results are not completely conclusive because of the correlation with OS, DFS, or LNM mainly depending on cut-off levels employed (197,198). Nevertheless, the utility of targeting two genes whose interaction is determining cell viability is a strategy that could be employed in the design of therapeutic options (synthetic lethality). The p53 and retinoblastoma protein deficiencies or the increased expression of Gli1 and SETD2 are cellular targets that could be employed for synthetic lethality in cervical cancer treatment (199-201).
The application of predictive markers in the clinical field faces two main challenges. On the one hand, there is the technical performance of the test; the sensitivity, specificity and cut-off value of the test are determining factors for its clinical interpretation. There is no homogenous procedure for developing, evaluating and reporting biomarker predictive performance. Multiple guidelines currently employed are designed to provide a generalized strategy for evaluating the quality and design of the protocol study according to pathological conditions (202). However, these guidelines in some cases provide little information about the study characteristics, which limits the reproducibility and the advance into the establishment of predictive performance. On the other hand, there is accessibility and coverage of healthcare programs, where the limited healthcare budget for CC treatment, reduces the availability for participating in therapeutic approaches based on precision medicine.
7. Conclusions
CC is largely preventable due to effective screening and vaccination programs. However, the vaccination rates remain low and only a minority of women can gain access to efficient screening programs. Therefore, CC-related mortality remains high in low-and middle-income countries. The clinical stage traditionally defines the treatment in CC patients; however, the sensitivity to treatment differs between individuals, even among those at the same stage. Despite extensive research in recent decades on the possible role of biological molecules such as proteins, DNA and non-coding RNA as predictive biomarkers, the identification of valid and reproducible response marker treatment is neither concise nor clear in CC because the data produced by the clinical studies are contradictory or not comparable due to the varied criteria used in the case selection in the clinical trials. Therefore, clinical studies with a larger number of patients and with clearly defined inclusion criteria are required to facilitate the integration of the information generated in each investigation. Moreover, the relevant findings must also be made accessible to the majority of patients.
Availability of data and materials
Not applicable.
Authors' contributions
ML, AC-P, ED-H, LC-M and AC-G performed the bibliographic review, wrote and critically revised the manuscript. ML and AC-P conceived and directed the present study. Data sharing is not applicable. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Authors' information
ORCID: Marcela Lizano, 0000-0002-7553-2541. Adriana Contreras-Paredes, 0000-0001-9965-4108. Erick De la Cruz-Hernández, 0000-0001-5236-7347.
Acknowledgments
Not applicable.
Funding
The present study was partially supported by CONAHCyT (grants no. CF-2019-263979 and PRONAII-7 Virus y Cáncer-303044); Instituto Nacional de Cancerología, México (grants no. 015/039/IBI/CEI/998/15 and 018/051/IBI/CEI/1294/18); Consejo Estatal de Ciencia y Tecnologia del Estado de Tabasco (grant no. PRODECTI-2023-01/090).
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
The International Agency for Research on Cancer: GLOBOCAN 2020: New Global Cancer Data. https://www.uicc.org/news/globocan-2020-new-global-cancer-data. Accessed December 17 2020 | |
|
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J and Bray F: Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob Health. 8:e191–203. 2020. View Article : Google Scholar : | |
|
Falcaro M, Castañon A, Ndlela B, Checchi M, Soldan K, Lopez-Bernal J, Elliss-Brookes L and Sasieni P: The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: A register-based observational study. Lancet. 398:2084–2092. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Moscicki AB, Schiffman M, Burchell A, Albero G, Giuliano AR, Goodman MT, Kjaer SK and Palefsky J: Updating the natural history of human papillomavirus and anogenital cancers. Vaccine. 30(Suppl 5): F24–F33. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Tommasino M: The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 26:13–21. 2014. View Article : Google Scholar | |
|
Hammer A, Rositch A, Qeadan F, Gravitt PE and Blaakaer J: Age-specific prevalence of HPV16/18 genotypes in cervical cancer: A systematic review and meta-analysis. Int J Cancer. 138:2795–2803. 2016. View Article : Google Scholar | |
|
Wright JD, Matsuo K, Huang Y, Tergas AI, Hou JY, Khoury-Collado F, St Clair CM, Ananth CV, Neugut AI and Hershman DL: Prognostic performance of the 2018 international federation of gynecology and obstetrics cervical cancer staging guidelines. Obstet Gynecol. 134:49–57. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
de Juan A, Redondo A, Rubio MJ, García Y, Cueva J, Gaba L, Yubero A, Alarcón J, Maximiano C and Oaknin A: SEOM clinical guidelines for cervical cancer (2019). Clin Transl Oncol. 22:270–278. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu SC, Huang EY, Hu CF, Ou YC, ChangChien CC, Wang CJ, Tsai CC, Fu HC, Wu CH and Lin H: Pretreatment factors associated with recurrence for patients with cervical cancer international federation of gynecology and obstetrics stage IB1 disease. Gynecol Obstet Invest. 81:339–345. 2016. View Article : Google Scholar | |
|
Rodriguez NM: Participatory innovation for human papillomavirus screening to accelerate the elimination of cervical cancer. Lancet Glob Health. 9:e582–e583. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou J, Lei N, Tian W, Guo R, Chen M, Qiu L, Wu F, Li Y and Chang L: Recent progress of the tumor microenvironmental metabolism in cervical cancer radioresistance. Front Oncol. 12:9996432022. View Article : Google Scholar : PubMed/NCBI | |
|
Yang J, Cai H, Xiao ZX, Wang H and Yang P: Effect of radiotherapy on the survival of cervical cancer patients: An analysis based on SEER database. Medicine (Baltimore). 98:e164212019. View Article : Google Scholar : PubMed/NCBI | |
|
U.S. Food and Drug Administration: About Biomarkers and Qualification. https://www.fda.gov/drugs/biomarker-qualificationprogram/about-biomarkers-and-qualification. Accessed July 7, 2021 | |
|
Volkova LV, Pashov AI and Omelchuk NN: Cervical Carcinoma: Oncobiology and Biomarkers. Int J Mol Sci. 22:125712021. View Article : Google Scholar : PubMed/NCBI | |
|
Ballman KV: Biomarker: Predictive or Prognostic? J Clin Oncol. 33:3968–3971. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Yusufaly TI, Zou J, Nelson TJ, Williamson CW, Simon A, Singhal M, Liu H, Wong H, Saenz CC, Mayadev J, et al: Improved Prognosis of Treatment Failure in Cervical Cancer with Nontumor PET/CT Radiomics. J Nucl Med. 63:1087–1093. 2022. View Article : Google Scholar : | |
|
Chang R, Qi S, Yue Y, Zhang X, Song J and Qian W: Predictive radiomic models for the chemotherapy response in non-small-cell lung cancer based on computerized-tomography images. Front Oncol. 11:6461902021. View Article : Google Scholar : PubMed/NCBI | |
|
Dhama K, Latheef SK, Dadar M, Samad HA, Munjal A, Khandia R, Karthik K, Tiwari R, Yatoo MI, Bhatt P, et al: Biomarkers in stress related diseases/disorders: diagnostic, prognostic, and therapeutic values. Front Mol Biosci. 6:912019. View Article : Google Scholar : PubMed/NCBI | |
|
Wang LH, Wu CF, Rajasekaran N and Shin YK: Loss of tumor suppressor gene function in human cancer: An overview. Cell Physiol Biochem. 51:2647–2693. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Tornesello ML, Faraonio R, Buonaguro L, Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and Buonaguro FM: The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front Oncol. 10:1502020. View Article : Google Scholar : PubMed/NCBI | |
|
Gyparaki MT, Basdra EK and Papavassiliou AG: DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Med (Berl). 91:1249–1256. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Charakorn C, Thadanipon K, Chaijindaratana S, Rattanasiri S, Numthavaj P and Thakkinstian A: The association between serum squamous cell carcinoma antigen and recurrence and survival of patients with cervical squamous cell carcinoma: A systematic review and meta-analysis. Gynecol Oncol. 150:190–200. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Dixit CK, Kadimisetty K, Otieno BA, Tang C, Malla S, Krause CE and Rusling JF: Electrochemistry-based approaches to low cost, high sensitivity, automated, multiplexed protein immunoassays for cancer diagnostics. Analyst. 141:536–547. 2016. View Article : Google Scholar : | |
|
Füzéry AK, Levin J, Chan MM and Chan DW: Translation of proteomic biomarkers into FDA approved cancer diagnostics: Issues and challenges. Clin Proteomics. 10:132013. View Article : Google Scholar : PubMed/NCBI | |
|
Sun Z, Shi Y, Shen Y, Cao L, Zhang W and Guan X: Analysis of different HER-2 mutations in breast cancer progression and drug resistance. J Cell Mol Med. 19:2691–2701. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Y, Zhang H, Zhang M, Meng Q, Cai L and Zhang Q: Elevation of serum CEA and CA15-3 levels during antitumor therapy predicts poor therapeutic response in advanced breast cancer patients. Oncol Lett. 14:7549–7556. 2017. | |
|
Alegría-Baños JA, Jiménez-López JC, Vergara-Castañeda A, de León DFC, Mohar-Betancourt A, Pérez-Montiel D, Sánchez-Domínguez G, García-Villarejo M, Olivares-Pérez C, Hernández-Constantino Á, et al: Kinetics of HE4 and CA125 as prognosis biomarkers during neoadjuvant chemotherapy in advanced epithelial ovarian cancer. J Ovarian Res. 14:962021. View Article : Google Scholar : PubMed/NCBI | |
|
Islam MS, Afrin S, Jones SI and Segars J: Selective progesterone receptor modulators-mechanisms and therapeutic utility. Endocr Rev. 41:bnaa0122020. View Article : Google Scholar : PubMed/NCBI | |
|
Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E and Cardoso F: Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer. 75:284–298. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Chimento A, De Luca A, Avena P, De Amicis F, Casaburi I, Sirianni R and Pezzi V: Estrogen receptors-mediated apoptosis in hormone-dependent cancers. Int J Mol Sci. 23:12422022. View Article : Google Scholar : PubMed/NCBI | |
|
Seale KN and Tkaczuk KHR: Circulating biomarkers in breast cancer. Clin Breast Cancer. 22:e319–e331. 2022. View Article : Google Scholar | |
|
Banerjee S, Yoon H, Ting S, Tang CM, Yebra M, Wenzel AT, Yeerna H, Mesirov JP, Wechsler-Reya RJ, Tamayo P and Sicklick JK: KITlow cells mediate imatinib resistance in gastrointestinal stromal tumor. Mol Cancer Ther. 20:2035–2048. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Jing W, Zhang R, Chen X, Zhang X and Qiu J: Association of glycosylation-related genes with different patterns of immune profiles and prognosis in cervical cancer. J Pers Med. 13:5292023. View Article : Google Scholar : PubMed/NCBI | |
|
National center for Biotechnology Information. https://clinicaltrials.gov/. 2023, Clinicaltrials.gov. | |
|
Hishinuma E, Shimada M, Matsukawa N, Li B, Motoike IN, Hagihara T, Shigeta S, Tokunaga H, Saigusa D, Kinoshita K, et al: Identification of predictive biomarkers for diagnosis and radiation sensitivity of uterine cervical cancer using wide-targeted metabolomics. J Obstet Gynaecol Res. 49:2109–2117. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Kilic S, Cracchiolo B, Gabel M, Haffty B and Mahmoud O: The relevance of molecular biomarkers in cervical cancer patients treated with radiotherapy. Ann Transl Med. 3:2612015.PubMed/NCBI | |
|
Rashid M, Zadeh LR, Baradaran B, Molavi O, Ghesmati Z, Sabzichi M and Ramezani F: Up-down regulation of HIF-1α in cancer progression. Gene. 798:1457962021. View Article : Google Scholar | |
|
Le QT and Courter D: Clinical biomarkers for hypoxia targeting. Cancer Metastasis Rev. 27:351–362. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Bishop AJ, Allen PK, Klopp AH, Meyer LA and Eifel PJ: Relationship Between Low Hemoglobin Levels and Outcomes After Treatment With Radiation or Chemoradiation in Patients With Cervical Cancer: Has the Impact of Anemia Been Overstated? Int J Radiat Oncol Biol Phys. 91:196–205. 2015. View Article : Google Scholar | |
|
Airley RE, Loncaster J, Raleigh JA, Harris AL, Davidson SE, Hunter RD, West CM and Stratford IJ: GLUT-1 and CAIX as intrinsic markers of hypoxia in carcinoma of the cervix: Relationship to pimonidazole binding. Int J Cancer. 104:85–91. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Feldser D, Agani F, Iyer NV, Pak B, Ferreira G and Semenza GL: Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 59:3915–3938. 1999.PubMed/NCBI | |
|
Domènech M, Hernández A, Plaja A, Martínez-Balibrea E and Balañà C: Hypoxia: The cornerstone of glioblastoma. Int J Mol Sci. 22:126082021. View Article : Google Scholar : PubMed/NCBI | |
|
Lei R, Li J, Liu F, Li W, Zhang S, Wang Y, Chu X and Xu J: HIF-1α promotes the keloid development through the activation of TGF-beta/Smad and TLR4/MyD88/NF-kappaB pathways. Cell Cycle. 18:3239–3250. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang PC, Liu X, Li MM, Ma YY, Sun HT, Tian XY, Wang Y, Liu M, Fu LS, Wang YF, et al: AT-533, a novel Hsp90 inhibitor, inhibits breast cancer growth and HIF-1α/VEGF/VEGFR-2-mediated angiogenesis in vitro and in vivo. Biochem Pharmacol. 172:1137712020. View Article : Google Scholar | |
|
Apte RS, Chen DS and Ferrara N: VEGF in signaling and disease: Beyond discovery and development. Cell. 176:1248–1264. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Yoshida K, Suzuki S, Sakata J, Utsumi F, Niimi K, Yoshikawa N, Nishino K, Shibata K, Kikkawa F and Kajiyama H: The upregulated expression of vascular endothelial growth factor in surgically treated patients with recurrent/radioresistant cervical cancer of the uterus. Oncol Lett. 16:515–521. 2018.PubMed/NCBI | |
|
Yan B, Ma QF, Tan WF, Cai HN, Li YL, Zhou ZG, Dai X, Zhu FX, Xiong YJ, Xu M, et al: Expression of HIF-1α is a predictive marker of the efficacy of neoadjuvant chemotherapy for locally advanced cervical cancer. Oncol Lett. 20:841–849. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Hu X, Xing L, Wei X, Liu X, Pang R, Qi L and Song S: Nonangiogenic function of VEGF and enhanced radiosensitivity of HeLa cells by inhibition of VEGF expression. Oncol Res. 20:93–101. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu P, Ou Y, Dong Y, Xu P and Yuan L: Expression of VEGF and HIF-1α in locally advanced cervical cancer: potential biomarkers for predicting preoperative radiochemotherapy sensitivity and prognosis. Onco Targets Ther. 9:3031–3037. 2016. | |
|
Wei LC, Wang N, Shi M, Liu JY, Li JP, Zhang Y, Huang YH, Li X and Chen Y: Clinical outcome observation of preoperative concurrent chemoradiotherapy/radiotherapy alone in 174 Chinese patients with local advanced cervical carcinoma. Onco Targets Ther. 6:67–74. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ferrandina G, Legge F, Fagotti A, Fanfani F, Distefano M, Morganti A, Cellini N and Scambia G: Preoperative concomitant chemoradiotherapy in locally advanced cervical cancer: Safety, outcome, and prognostic measures. Gynecol Oncol. 107(1 Suppl 1): S127–S132. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Lèguevaque P, Motton S, Delannes M, Querleu D, Soulé-Tholy M, Tap G and Houvenaeghel G: Completion surgery or not after concurrent chemoradiotherapy for locally advanced cervical cancer? Eur J Obstet Gynecol Reprod Biol. 155:188–192. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Sharma M, Khan R, Aggarwal M and Sharma A: Modulatory effects of chemoradiation on angiogenic factors and laminin in cervical cancer: Link with treatment response. Asian Pac J Cancer Prev. 18:2937–2944. 2017.PubMed/NCBI | |
|
Nguyen VT, Winterman S, Playe M, Benbara A, Zelek L, Pamoukdjian F and Bousquet G: Dose-Intense cisplatin-based neoadjuvant chemotherapy increases survival in advanced cervical cancer: An up-to-date meta-analysis. Cancers (Basel). 14:8422022. View Article : Google Scholar : PubMed/NCBI | |
|
Angioli R, Plotti F, Montera R, Aloisi A, Luvero D, Capriglione S, Terranova C, De Cicco Nardone C, Muzii L and Benedetti-Panici P: Neoadjuvant chemotherapy plus radical surgery followed by chemotherapy in locally advanced cervical cancer. Gynecol Oncol. 127:290–296. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao K, Hu M, Yang R, Liu J, Zeng P and Zhao T: Decreasing expression of HIF-1α, VEGF-A, and Ki67 with efficacy of neoadjuvant therapy in locally advanced cervical cancer. Medicine (Baltimore). 102:e338202023. View Article : Google Scholar | |
|
Hu Y, Han Y, Shen Y, Chen J, Chen Y, Chen Y, Tang J, Xue M, Hong L, Cheng W, et al: Neoadjuvant chemotherapy for patients with international federation of gynecology and obstetrics stages IB3 and IIA2 cervical cancer: A multicenter prospective trial. BMC Cancer. 202:12702022. View Article : Google Scholar | |
|
Datta A, West C, O'Connor JPB, Choudhury A and Hoskin P: Impact of hypoxia on cervical cancer outcomes. Int J Gynecol Cancer. 31:1459–1470. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Dunst J, Kuhnt T, Strauss HG, Krause U, Pelz T, Koelbl H and Haensgen G: Anemia in cervical cancers: Impact on survival, patterns of relapse, and association with hypoxia and angiogenesis. Int J Radiat Oncol Biol Phys. 56:778–787. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Shin NR, Lee YY, Kim SH, Choi CH, Kim TJ, Lee JW, Bae DS and Kim BG: Prognostic value of pretreatment hemoglobin level in patients with early cervical cancer. Obstet Gynecol Sci. 57:28–36. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Barkati M, Fortin I, Mileshkin L, Bernshaw D, Carrier JF and Narayan K: Hemoglobin level in cervical cancer: a surrogate for an infiltrative phenotype. Int J Gynecol Cancer. 23:724–729. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Gennigens C, De Cuypere M, Seidel L, Hermesse J, Barbeaux A, Forget F, Albert A, Jerusalem G and Kridelka F: Correlation between hematological parameters and outcome in patients with locally advanced cervical cancer treated by concomitant chemoradiotherapy. Cancer Med. 9:8432–8443. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Moreno-Acosta P, Carrillo S, Gamboa O, Romero-Rojas A, Acosta J, Molano M, Balart-Serra J, Cotes M, Rancoule C and Magné N: Novel predictive biomarkers for cervical cancer prognosis. Mol Clin Oncol. 5:792–796. 2016. View Article : Google Scholar | |
|
Kasi PM and Grothey A: Chemotherapy-Induced neutropenia as a prognostic and predictive marker of outcomes in solid-tumor patients. Drugs. 78:737–745. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Markovina S, Wang S, Henke LE, Luke CJ, Pak SC, DeWees T, Pfeifer JD, Schwarz JK, Liu W, Chen S, et al: Serum squamous cell carcinoma antigen as an early indicator of response during therapy of cervical cancer. Br J Cancer. 118:72–78. 2018. View Article : Google Scholar : | |
|
Chen W, Xiu S, Xie X, Guo H, Xu Y, Bai P and Xia X: Prognostic value of tumor measurement parameters and SCC-Ag changes in patients with locally-advanced cervical cancer. Radiat Oncol. 17:62022. View Article : Google Scholar : PubMed/NCBI | |
|
Choi KH, Lee SW, Yu M, Jeong S, Lee JW and Lee JH: Significance of elevated SCC-Ag level on tumor recurrence and patient survival in patients with squamous-cell carcinoma of uterine cervix following definitive chemoradiotherapy: A multi-institutional analysis. J Gynecol Oncol. 30:e12019. View Article : Google Scholar | |
|
Xu D, Wang D, Wang S, Tian Y, Long Z and Ren X: Correlation between squamous cell carcinoma antigen level and the clinicopathological features of early-stage cervical squamous cell carcinoma and the predictive value of squamous cell carcinoma antigen combined with computed tomography scan for lymph node metastasis. Int J Gynecol Cancer. 27:1935–1942. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Tantari M, Bogliolo S, Morotti M, Balaya V, Bouttitie F, Buenerd A, Magaud L, Lecuru F, Guani B, Mathevet P, et al: Lymph node involvement in early-stage cervical cancer: Is lymphangiogenesis a risk factor? Results from the MICROCOL Study. Cancers (Basel). 14:2122022. View Article : Google Scholar : PubMed/NCBI | |
|
Kang S, Nam BH, Park JY, Seo SS, Ryu SY, Kim JW, Kim SC, Park SY and Nam JH: Risk Assessment tool for distant recurrence after platinum-based concurrent chemoradiation in patients with locally advanced cervical cancer: A Korean gynecologic oncology group study. J Clin Oncol. 30:2369–2374. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Mabuchi S, Isohashi F, Yokoi T, Takemura M, Yoshino K, Shiki Y, Ito K, Enomoto T, Ogawa K and Kimura T: A phase II study of postoperative concurrent carboplatin and paclitaxel combined with intensity-modulated pelvic radiotherapy followed by consolidation chemotherapy in surgically treated cervical cancer patients with positive pelvic lymph nodes. Gynecol Oncol. 141:240–246. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Shim SH, Kim DY, Lee SJ, Kim SN, Kang SB, Lee SW, Park JY, Suh DS, Kim JH, Kim YM, et al: Prediction model for para-aortic lymph node metastasis in patients with locally advanced cervical cancer. Gynecol Oncol. 144:40–45. 2017. View Article : Google Scholar | |
|
Yoon HI, Cha J, Keum KC, Lee HY, Nam EJ, Kim SW, Kim S, Kim YT, Kim GE and Kim YB: Treatment outcomes of extended-field radiation therapy and the effect of concurrent chemotherapy on uterine cervical cancer with para-aortic lymph node metastasis. Radiat Oncol. 10:182015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang G, Fu C, Zhang Y, Wang J, Qiao N, Yang Q and Cheng Y: Extended-Field Intensity-Modulated Radiotherapy and Concurrent Cisplatin-Based Chemotherapy for Postoperative Cervical Cancer With Common Iliac or Para-Aortic Lymph Node Metastases: A Retrospective Review in a Single Institution. Int J Gynecol Cancer. 22:1220–1225. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Walker JL, Morrison A, DiSilvestro P and von Gruenigen VE; Gynecologic Oncology Group: A phase I/II study of extended field radiation therapy with concomitant paclitaxel and cisplatin chemotherapy in patients with cervical carcinoma metastatic to the para-aortic lymph nodes: A gynecologic oncology group study. Gynecol Oncol. 112:78–84. 2009. View Article : Google Scholar | |
|
Matsuo K, Shimada M, Aoki Y, Sakamoto M, Takeshima N, Fujiwara H, Matsumoto T, Mikami M and Sugiyama T: Comparison of adjuvant therapy for node-positive clinical stage IB-IIB cervical cancer: Systemic chemotherapy versus pelvic irradiation. Int J Cancer. 141:1042–1051. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Jung PS, Kim DY, Lee SW, Park JY, Suh DS, Kim JH, Kim YM, Kim YT and Nam JH: clinical role of adjuvant chemotherapy after radical hysterectomy for FIGO Stage IB-IIA cervical cancer: Comparison with Adjuvant RT/CCRT using inverse-probability-of-treatment weighting. PLoS One. 10:e01322982015. View Article : Google Scholar : PubMed/NCBI | |
|
Strauss HG, Laban C, Lautenschläger C, Buchmann J, Schneider I and Koelbl H: SCC antigen in the serum as an independent prognostic factor in operable squamous cell carcinoma of the cervix. Eur J Cancer. 38:1987–1991. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang G, Miao L, Wu H, Zhang Y and Fu C: Pretreatment squamous cell carcinoma antigen (SCC-Ag) as a predictive factor for the use of consolidation chemotherapy in cervical cancer patients after postoperative extended-field concurrent chemoradiotherapy. Technol Cancer Res Treat. 20:153303382110446262021. View Article : Google Scholar : PubMed/NCBI | |
|
Kawaguchi R, Furukawa N, Kobayashi H and Asakawa I: Posttreatment cut-off levels of squamous cell carcinoma antigen as a prognostic factor in patients with locally advanced cervical cancer treated with radiotherapy. J Gynecol Oncol. 24:313–320. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ryu HK, Baek JS, Kang WD and Kim SM: The prognostic value of squamous cell carcinoma antigen for predicting tumor recurrence in cervical squamous cell carcinoma patients. Obstet Gynecol Sci. 58:368–376. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Gadducci A, Tana R, Cosio S and Genazzani AR: The serum assay of tumour markers in the prognostic evaluation, treatment monitoring and follow-up of patients with cervical cancer: A review of the literature. Crit Rev Oncol Hematol. 66:10–20. 2008. View Article : Google Scholar | |
|
Jiang M, Chen P, Guo X, Zhang X, Gao Q, Zhang J, Zhao G and Zheng J: Identification of EGFR mutation status in male patients with non-small-cell lung cancer: Role of 18F-FDG PET/CT and serum tumor markers CYFRA21-1 and SCC-Ag. EJNMMI Res. 13:272023. View Article : Google Scholar : | |
|
Liu Z and Shi H: Prognostic role of squamous cell carcinoma antigen in cervical cancer: A Meta-analysis. Dis Markers. 2019:67103522019. View Article : Google Scholar : PubMed/NCBI | |
|
Anastasi E, Gigli S, Ballesio L, Angeloni A and Manganaro L: The complementary role of imaging and tumor biomarkers in gynecological cancers: An update of the literature. Asian Pac J Cancer Prev. 19:309–317. 2018.PubMed/NCBI | |
|
Cortés-Ciriano I, Lee JJK, Xi R, Jain D, Jung YL, Yang L, Gordenin D, Klimczak LJ, Zhang CZ, Pellman DS, et al: Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 52:331–341. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Müller D and Győrffy B: DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim Biophys Acta Rev Cancer. 1877:1887222022. View Article : Google Scholar : PubMed/NCBI | |
|
He L, Zhao C, Zhang Q, Zinta G, Wang D, Lozano-Durán R and Zhu JK: Pathway conversion enables a double-lock mechanism to maintain DNA methylation and genome stability. Proc Natl Acad Sci USA. 118:e21073201182021. View Article : Google Scholar : PubMed/NCBI | |
|
Portela A and Esteller M: Epigenetic modifications and human disease. Nat Biotechnol. 28:1057–1068. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Vural S, Palmisano A, Reinhold WC, Pommier Y, Teicher BA and Krushkal J: Association of expression of epigenetic molecular factors with DNA methylation and sensitivity to chemotherapeutic agents in cancer cell lines. Clin Epigenetics. 13:492021. View Article : Google Scholar : PubMed/NCBI | |
|
Xing W, Sun H, Yan C, Zhao C, Wang D, Li M and Ma J: A prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant from benign pulmonary nodules. BMC Cancer. 21:2632021. View Article : Google Scholar : PubMed/NCBI | |
|
Levenson VV: DNA methylation as a universal biomarker. Expert Rev Mol Diagn. 10:481–488. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC and Ross JP: DNA Methylation cancer biomarkers: Translation to the Clinic. Front Genet. 10:11502019. View Article : Google Scholar : PubMed/NCBI | |
|
Clarke MA, Luhn P, Gage JC, Bodelon C, Dunn ST, Walker J, Zuna R, Hewitt S, Killian JK, Yan L, et al: Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int J Cancer. 141:701–710. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
El Aliani A, El-Abid H, El Mallali Y, Attaleb M, Ennaji MM and El Mzibri M: Association between Gene Promoter Methylation and Cervical Cancer Development: Global Distribution and A Meta-analysis. Cancer Epidemiol Biomarkers Prev. 30:450–459. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu J, Nie S, Li S, Meng H, Sun R, Yang J and Cheng W: Methylation-driven genes and their prognostic value in cervical squamous cell carcinoma. Ann Transl Med. 8:868. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Salta S, Maia-Moço L, Estevão-Pereira H, Sequeira JP, Vieira R, Bartosch C, Petronilho S, Monteiro P, Sousa A, Baldaque I, et al: Performance of DNA methylation-based biomarkers in the cervical cancer screening program of northern Portugal: A feasibility study. Int J Cancer. 149:1916–1925. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Verhoef VM, Bosgraaf RP, van Kemenade FJ, Rozendaal L, Heideman DAM, Hesselink AT, Bekkers RL, Steenbergen RD, Massuger LF, Melchers WJ, et al: Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): A randomised controlled non-inferiority trial. Lancet Oncol. 15:315–322. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Sood S, Patel FD, Ghosh S, Arora A, Dhaliwal LK and Srinivasan R: Epigenetic Alteration by DNA Methylation of ESR1, MYOD1 and hTERT gene promoters is useful for prediction of response in patients of locally advanced invasive cervical carcinoma treated by chemoradiation. Clin Oncol (R Coll Radiol). 27:720–727. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Contreras-Romero C, Pérez-Yépez EA, Martinez-Gutierrez AD, Campos-Parra A, Zentella-Dehesa A, Jacobo-Herrera N, López-Camarillo C, Corredor-Alonso G, Martínez-Coronel J, Rodríguez-Dorantes M, et al: Gene Promoter-Methylation signature as biomarker to predict cisplatin-radiotherapy sensitivity in locally advanced cervical cancer. Front Oncol. 12:7734382022. View Article : Google Scholar : PubMed/NCBI | |
|
Guerrero-Setas D, Pérez-Janices N, Blanco-Fernandez L, Ojer A, Cambra K, Berdasco M, Esteller M, Maria-Ruiz S, Torrea N and Guarch R: RASSF2 hypermethylation is present and related to shorter survival in squamous cervical cancer. Mod Pathol. 26:1111–1122. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Yamazaki Y, Ur rutia R, Franco LM, Giliani S, Zhang K, Alazami AM, Dobbs AK, Masneri S, Joshi A, Otaizo-Carrasquero F, et al: PAX1 is essential for development and function of the human thymus. Sci Immunol. 5:eaax10362020. View Article : Google Scholar : PubMed/NCBI | |
|
Wu W, Kong X, Jia Y, Jia Y, Ou W, Dai C, Li G and Gao R: An overview of PAX1: Expression, function and regulation in development and diseases. Front Cell Dev Biol. 10:10511022022. View Article : Google Scholar : PubMed/NCBI | |
|
Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI, Chan MWY, Liao CC, Chen CW, Lin WY and Chang CC: Paired Box-1 (PAX1) activates multiple phosphatases and inhibits kinase cascades in cervical cancer. Sci Rep. 9:91952019. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Zhou X, Zeng M, Zhou Y, Zhang Y, Liou YL and Zhu H: Methylation of PAX1 gene promoter in the prediction of concurrent chemo-radiotherapy efficacy in cervical cancer. J Cancer. 12:5136–5143. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Yang W, Zhang Z, Li L, Zhang K, Xu Y, Xia M, Zhou J, Gong Y, Chen J and Gong K: ZNF582 overexpression restrains the progression of clear cell renal cell carcinoma by enhancing the binding of TJP2 and ERK2 and inhibiting ERK2 phosphorylation. Cell Death Dis. 14:2122023. View Article : Google Scholar : PubMed/NCBI | |
|
Li N, He Y, Mi P and Hu Y: ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: A meta-analysis of related studies in Chinese population. Medicine (Baltimore). 98:e142972019. View Article : Google Scholar : PubMed/NCBI | |
|
Huang J, Wang G, Tang J, Zhuang W, Wang LP, Liou YL, Liu YZ, Zhou HH and Zhu YS: DNA Methylation Status of PAX1 and ZNF582 in esophageal squamous cell carcinoma. Int J Environ Res Public Health. 14:2162017. View Article : Google Scholar : PubMed/NCBI | |
|
Huang RL, Chang CC, Su PH, Chen YC, Liao YP, Wang HC, Yo YT, Chao TK, Huang HC, Lin CY, et al: Methylomic Analysis Identifies Frequent DNA Methylation of Zinc Finger Protein 582 (ZNF582) in Cervical Neoplasms. PLoS One. 7:e410602012. View Article : Google Scholar : PubMed/NCBI | |
|
Wu NYY, Zhang X, Chu T, Zhu S, Deng Y, Zhou Y, Wang Y, Zhao X, Liu L, Fang C, et al: High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med. 7:3282019. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan T, Edelmann D, Fan Z, Alwers E, Kather JN, Brenner H and Hoffmeister M: Machine learning in the identification of prognostic DNA methylation biomarkers among patients with cancer: A systematic review of epigenome-wide studies. Artif Intell Med. 143:1025892023. View Article : Google Scholar : PubMed/NCBI | |
|
Ma JH, Huang Y, Liu LY and Feng Z: An 8-gene DNA methylation signature predicts the recurrence risk of cervical cancer. J Int Med Res. 49:030006052110184432021. View Article : Google Scholar : PubMed/NCBI | |
|
Cech TR and Steitz JA: The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 157:77–94. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Weng Q, Ge J, Zhang X, Guo J and Ye G: tRNA-derived small RNAs: Mechanisms and potential roles in cancers. Genes Dis. 9:1431–1442. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S, et al: PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol. 23:424–436. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Mendell JT: MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang J, Chen B, Gan C, Sun H, Zhang J and Feng L: A Comprehensive Review of Small Interfering RNAs (siRNAs): Mechanism, therapeutic targets, and delivery strategies for cancer therapy. Int J Nanomedicine. 18:7605–7635. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Matera AG, Terns RM and Terns MP: Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 8:209–220. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and Chen M: Circular RNA: An emerging key player in RNA world. Brief Bioinform. 18:547–557. 2017. | |
|
Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME, Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Fort V, Khelifi G and Hussein SMI: Long non-coding RNAs and transposable elements: A functional relationship. Biochim Biophys Acta Mol Cell Res. 1868:1188372021. View Article : Google Scholar | |
|
Ozata DM, Gainetdinov I, Zoch A, O'Carroll D and Zamore PD: PIWI-interacting RNAs: Small RNAs with big functions. Nat Rev Genet. 20:89–108. 2019. View Article : Google Scholar | |
|
Abramowicz A and Story MD: The long and short of It: The emerging roles of non-coding RNA in small extracellular vesicles. Cancers (Basel). 12:14452020. View Article : Google Scholar : PubMed/NCBI | |
|
Hahne JC, Lampis A and Valeri N: Vault RNAs: Hidden gems in RNA and protein regulation. Cell Mol Life Sci. 78:1487–1499. 2021. View Article : Google Scholar : | |
|
Fan CM, Wang JP, Tang YY, Zhao J, He SY, Xiong F, Guo C, Xiang B, Zhou M, Li XL, et al: circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 110:2180–2188. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Toden S, Zumwalt TJ and Goel A: Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 1875:1884912021. View Article : Google Scholar : | |
|
Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C, Guo C, Li X, Liao Q, Zhang W, et al: Upregulation and hypomethylation of lncRNA AFAP1-AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 41:2431–2439. 2019.PubMed/NCBI | |
|
Xie M, Ma L, Xu T, Pan Y, Wang Q, Wei Y and Shu Y: Potential regulatory roles of MicroRNAs and Long Noncoding RNAs in Anticancer Therapies. Mol Ther Nucleic Acids. 13:233–243. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Lampropoulou DI, Pliakou E, Aravantinos G, Filippou D and Gazouli M: The Role of Exosomal Non-Coding RNAs in colorectal cancer drug resistance. Int J Mol Sci. 23:14732022. View Article : Google Scholar : PubMed/NCBI | |
|
Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al: MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 109:E2110–E2116. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R and Nishikura K: Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 13:13–21. 2006. View Article : Google Scholar | |
|
Zou K, Yang E, Cui T and Li Z: Circulating miR-326 could serve as a predictive biomarker for response to neoadjuvant chemotherapy in locally advanced cervical cancer. Front Oncol. 12:10367102022. View Article : Google Scholar : PubMed/NCBI | |
|
Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW and Wang X: A microRNA expression signature for cervical cancer prognosis. Cancer Res. 70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Pedroza-Torres A, Fernández-Retana J, Peralta-Zaragoza O, Jacobo-Herrera N, Cantú de Leon D, Cerna-Cortés JF, Lopez-Camarillo C and Pérez-Plasencia C: A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 142:557–565. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Q, Qin J, Chen A, Zhou J, Liu J, Cheng J, Qiu J and Zhang J: Downregulation of microRNA-145 is associated with aggressive progression and poor prognosis in human cervical cancer. Tumour Biol. 36:3703–3708. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Fekete JT, Welker Á and Győrffy B: miRNA expression signatures of therapy response in squamous cell carcinomas. Cancers (Basel). 13:632020. View Article : Google Scholar | |
|
Wei H, Wen-Ming C and Jun-Bo J: Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J Int Med Res. 45:1054–1060. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Shi M, Du L, Liu D, Qian L, Hu M, Yu M, Yang Z, Zhao M, Chen C, Guo L, et al: Glucocorticoid regulation of a novel HPV-E6-p53-miR-145 pathway modulates invasion and therapy resistance of cervical cancer cells. J Pathol. 228:148–157. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Liu M, An J, Huang M, Wang L, Tu B, Song Y, Ma K, Wang Y, Wang S, Zhu H, et al: MicroRNA-492 overexpression involves in cell proliferation, migration, and radiotherapy response of cervical squamous cell carcinomas. Mol Carcinog. 57:32–43. 2018. View Article : Google Scholar | |
|
Barik S, Mitra S, Suryavanshi M, Dewan A, Kaur I, Kumar D, Mishra M and Vishwakarma G: To study the role of pre-treatment microRNA (micro ribonucleic acid) expression as a predictor of response to chemoradiation in locally advanced carcinoma cervix. Cancer Rep. 4:e13482021. View Article : Google Scholar | |
|
Wei W and Liu C: Prognostic and predictive roles of microRNA-411 and its target STK17A in evaluating radiotherapy efficacy and their effects on cell migration and invasion via the p53 signaling pathway in cervical cancer. Mol Med Rep. 21:267–281. 2020. | |
|
Zuccherato LW, Machado CMT, Magalhães WCS, Martins PR, Campos LS, Braga LC, Teixeira-Carvalho A, Martins-Filho OA, Franco TMRF, Paula SOC, et al: Cervical Cancer Stem-Like Cell Transcriptome Profiles Predict Response to Chemoradiotherapy. Front Oncol. 11:6393392021. View Article : Google Scholar : PubMed/NCBI | |
|
Hasanzadeh M, Movahedi M, Rejali M, Maleki F, Moetamani-Ahmadi M, Seifi S, Hosseini Z, Khazaei M, Amerizadeh F, Ferns GA, et al: The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J Cell Physiol. 234:1289–1294. 2019. View Article : Google Scholar | |
|
Ravegnini G, Gorini F, Dondi G, Tesei M, De Crescenzo E, Morganti AG, Hrelia P, De Iaco P, Angelini S and Perrone AM: Emerging Role of MicroRNAs in the therapeutic response in cervical cancer: A systematic review. Front Oncol. 12:8479742022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu C and Wang R: The roles of hedgehog signaling pathway in radioresistance of cervical cancer. Dose Response. 17:15593258198852932019. View Article : Google Scholar : PubMed/NCBI | |
|
Fang F, Guo C, Zheng W, Wang Q and Zhou L: Exosome-Mediated Transfer of miR-1323 from Cancer-Associated Fibroblasts Confers Radioresistance of C33A Cells by Targeting PABPN1 and Activating Wnt/β-Catenin signaling pathway in cervical cancer. Reprod Sci. 29:1809–1821. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Che Y, Li Y, Zheng F, Zou K, Li Z, Chen M, Hu S, Tian C, Yu W, Guo W, et al: TRIP4 promotes tumor growth and metastasis and regulates radiosensitivity of cervical cancer by activating MAPK, PI3K/AKT, and hTERT signaling. Cancer Lett. 452:1–13. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wen X, Liu S, Sheng J and Cui M: Recent advances in the contribution of noncoding RNAs to cisplatin resistance in cervical cancer. PeerJ. 8:e92342020. View Article : Google Scholar : PubMed/NCBI | |
|
Sun Y, Feng Y, Zhang G and Xu Y: The endonuclease APE1 processes miR-92b formation, thereby regulating expression of the tumor suppressor LDLR in cervical cancer cells. Ther Adv Med Oncol. 11:17588359198558592019. View Article : Google Scholar : PubMed/NCBI | |
|
Yang F, Guo L, Cao Y, Li S, Li J and Liu M: MicroRNA-7-5p Promotes Cisplatin Resistance of Cervical Cancer Cells and Modulation of Cellular Energy Homeostasis by Regulating the Expression of the PARP-1 and BCL2 Genes. Med Sci Monit. 24:6506–6516. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
St Laurent G, Wahlestedt C and Kapranov P: The Landscape of long noncoding RNA classification. Trends Genet. 31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Luo F, Wen Y, Zhou H and Li Z: Roles of long non-coding RNAs in cervical cancer. Life Sci. 256:1179812020. View Article : Google Scholar : PubMed/NCBI | |
|
Chandra Gupta S and Nandan Tripathi Y: Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967. 2017. View Article : Google Scholar | |
|
Zhang P, Wu W, Chen Q and Chen M: Non-Coding RNAs and their Integrated Networks. J Integr Bioinform. 16:201900272019. View Article : Google Scholar : PubMed/NCBI | |
|
Ma L, Bajic VB and Zhang Z: On the classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang W, Wu Q, Liu Y, Wang X, Ma C and Zhu W: LncRNA HOTAIR promotes chemoresistance by facilitating epithelial to mesenchymal transition through miR-29b/PTEN/PI3K signaling in cervical cancer. Cells Tissues Organs. 211:16–29. 2022. View Article : Google Scholar | |
|
Zhou Y, Wang Y, Lin M, Wu D and Zhao M: LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell Int. 21:4002021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou M, Liu L, Wang J and Liu W: The role of long noncoding RNAs in therapeutic resistance in cervical cancer. Front Cell Dev Biol. 10:10609092022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Fang C, Feng Z, Xia T, Lu L, Luo M, Chen Y, Liu Y and Li Y: The Role of LncRNAs in the regulation of radiotherapy sensitivity in cervical cancer. Front Oncol. 12:8968402022. View Article : Google Scholar : PubMed/NCBI | |
|
Li N, Meng DD, Gao L, Xu Y, Liu PJ, Tian YW, Yi ZY, Zhang Y, Tie XJ and Xu ZQ: Overexpression of HOTAIR leads to radioresistance of human cervical cancer via promoting HIF-1α expression. Radiat Oncol. 13:2102018. View Article : Google Scholar | |
|
Liu S, Zhang M and Qu P: Expression level and clinical significance of HOX transcript antisense intergenic RNA in cervical cancer: A meta-analysis. Sci Rep. 6:380472016. View Article : Google Scholar : PubMed/NCBI | |
|
Lu H, He Y, Lin L, Qi Z, Ma L, Li L and Su Y: Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 37:1683–1691. 2016. View Article : Google Scholar | |
|
Gao J, Liu L, Li G, Cai M, Tan C, Han X and Han L: LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 126:994–1001. 2019. View Article : Google Scholar | |
|
Fang X, Zhong G, Wang Y, Lin Z, Lin R and Yao T: Low GAS5 expression may predict poor survival and cisplatin resistance in cervical cancer. Cell Death Dis. 11:5312020. View Article : Google Scholar : PubMed/NCBI | |
|
Ge X, Gu Y, Li D, Jiang M, Zhao S, Li Z and Liu S: Knockdown of lncRNA PCAT1 Enhances Radiosensitivity of Cervical Cancer by Regulating miR-128/GOLM1 Axis. Onco Targets Ther. 13:10373–10385. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Belousova EA, Filipenko ML and Kushlinskii NE: Circular RNA: New regulatory molecules. Bull Exp Biol Med. 164:803–815. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Chen LL and Yang L: Regulation of circRNA biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Guttman M, Russell P, Ingolia NT, Weissman JS and Lander ES: Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 154:240–251. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ: Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F, Nazarali AJ and Ji S: Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 6:1167–1176. 2016.PubMed/NCBI | |
|
Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H and Jiang Y: Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 19:1012020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou SY, Chen W, Yang SJ, Xu ZH, Hu JH, Zhang HD, Zhong SL and Tang JH: The emerging role of circular RNAs in breast cancer. Biosci Rep. 39:BSR201906212019. View Article : Google Scholar : PubMed/NCBI | |
|
Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, Ye Y, Zhang Z, Mills T, Feng J, et al: Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 11:552019. View Article : Google Scholar | |
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar : | |
|
Han B, Chao J and Yao H: Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Hussen BM, Honarmand Tamizkar K, Hidayat HJ, Taheri M and Ghafouri-Fard S: The role of circular RNAs in the development of hepatocellular carcinoma. Pathol Res Pract. 223:1534952021. View Article : Google Scholar : PubMed/NCBI | |
|
Pan G, Mao A, Liu J, Lu J, Ding J and Liu W: Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 53:e127202020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 19:1102020. View Article : Google Scholar : PubMed/NCBI | |
|
Sun G, Li Z, He Z, Wang W, Wang S, Zhang X, Cao J, Xu P, Wang H, Huang X, et al: Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J Exp Clin Cancer Res. 39:2462020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E and Gong C: Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 10:552019. View Article : Google Scholar : PubMed/NCBI | |
|
Luo YH, Yang YP, Chien CS, Yarmishyn AA, Ishola AA, Chien Y, Chen YM, Huang TW, Lee KY, Huang WC, et al: Plasma Level of Circular RNA hsa_circ_0000190 Correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers (Basel). 12:17402020. View Article : Google Scholar : PubMed/NCBI | |
|
Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC, Garmy-Susini B and Lacazette E: How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. 164:45–52. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X and Zheng X: Hsa_circ_0001495 contributes to cervical cancer progression by targeting miR-526b-3p/TMBIM6/mTOR axis. Reprod Biol. 22:1006482022. View Article : Google Scholar : PubMed/NCBI | |
|
Xie H, Wang J and Wang B: Circular RNA Circ_0003221 promotes cervical cancer progression by regulating miR-758-3p/CPEB4 Axis. Cancer Manag Res. 13:5337–5350. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X and Ding Y: Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids. 19:413–420. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wang YM, Huang LM, Li DR, Shao JH, Xiong SL, Wang CM and Lu SM: Hsa_circ_0101996 combined with hsa_circ_0101119 in peripheral whole blood can serve as the potential biomarkers for human cervical squamous cell carcinoma. Int J Clin Exp Pathol. 10:11924–11931. 2017.PubMed/NCBI | |
|
Li N, Liu J and Deng X: Identification of a novel circRNA, hsa_circ_0065898, that regulates tumor growth in cervical squamous cell carcinoma. Transl Cancer Res. 10:47–56. 2021. View Article : Google Scholar | |
|
Zhang X, Yang S, Chen W, Dong X, Zhang R, Ye H, Ye H, Mei X, Liu H, Fang Y and He C: Circular RNA circYPEL2: A novel biomarker in cervical cancer. Genes (Basel). 13:382021. View Article : Google Scholar | |
|
Wu P, Qin J, Liu L, Tan W, Lei L and Zhu J: circEPSTI1 promotes tumor progression and cisplatin resistance via upregulating MSH2 in cervical cancer. Aging (Albany NY). 14:5406–5416. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Y, Lan Y, Chi Y, Yang B and Ren C: Downregulation of Circ-CEP128 enhances the paclitaxel sensitivity of cervical cancer through regulating miR-432-5p/MCL1. Biochem Genet. 60:2346–2363. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Dong M, Li P, Xie Y, Wang Z and Wang R: CircMYBL2 regulates the resistance of cervical cancer cells to paclitaxel via miR -665-dependent regulation of EGFR. Drug Dev Res. 82:1193–1205. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao X, Dong W, Luo G, Xie J, Liu J and Yu F: Silencing of hsa_circ_0009035 suppresses cervical cancer progression and enhances radiosensitivity through MicroRNA 889-3p-Dependent regulation of HOXB7. Mol Cell Biol. 41:e00631202021. View Article : Google Scholar : PubMed/NCBI | |
|
Cho O, Chun M, Oh YT, Noh OK, Chang SJ, Ryu HS and Lee EJ: Prognostic implication of simultaneous anemia and lymphopenia during concurrent chemoradiotherapy in cervical squamous cell carcinoma. Tumour Biol. 39:10104283177331442017. View Article : Google Scholar : PubMed/NCBI | |
|
Leetanaporn K and Hanprasertpong J: Predictive value of the hemoglobin-albumin-lymphocyte-platelet (HALP) index on the oncological outcomes of locally advanced cervical cancer patients. Cancer Manag Res. 14:1961–1972. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Tang Z, Zeng M, Wang X, Guo C, Yue P, Zhang X, Lou H, Chen J, Mu D, Kong D, et al: Synthetic lethality between TP53 and ENDOD1. Nat Commun. 13:28612022. View Article : Google Scholar : PubMed/NCBI | |
|
Lyu J, Yang EJ, Zhang B, Wu C, Pardeshi L, Shi C, Mou PK, Liu Y, Tan K and Shim JS: Synthetic lethality of RB1 and aurora A is driven by stathmin-mediated disruption of microtubule dynamics. Nat Commun. 11:51052020. View Article : Google Scholar : PubMed/NCBI | |
|
Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, Mavrommati I, Pai CC, Zalmas LP, Drobnitzky N, et al: Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell. 28:557–568. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Sauerbrei W, Haeussler T, Balmford J and Huebner M: Structured reporting to improve transparency of analyses in prognostic marker studies. BMC Med. 20:1842022. View Article : Google Scholar : PubMed/NCBI |