CHCHD2 mediates glioblastoma cell proliferation, mitochondrial metabolism, hypoxia‑induced invasion and therapeutic resistance
- Authors:
- Published online on: August 25, 2023 https://doi.org/10.3892/ijo.2023.5565
- Article Number: 117
-
Copyright: © Lumibao et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Glioblastoma (GBM; WHO grade IV glioma) is the most common, malignant and aggressive form of primary brain tumor affecting adults, accounting for ~50% of diagnosed gliomas each year (1). Patients with GBM present with a median survival time of only 15-20 months, with only 5-10% of patients surviving after 5 years (2). Despite the current multimodal standard of care, which consists of maximal surgical resection followed by radiotherapy and chemotherapy with the DNA alkylating agent, temozolomide (TMZ), the overall prognosis of patients remains poor, underscoring the need for a more in-depth understanding of tumor biology to inspire the identification of novel therapeutic targets. Contributing to tumor aggressiveness and universal recurrence are GBM hallmarks, including but not limited to: Rapid, diffuse invasion into the surrounding brain parenchyma, substantial chemo- and radioresistance and rapid adaptation to microenvironmental stressors, such as hypoxia (3,4).
The amplification of the epidermal growth factor receptor (EGFR), one of the most common genetic abnormalities observed in GBM tumors (5), leads to increased proliferative and anti-apoptotic signaling, as well as invasive behavior (6,7). Tumors that exhibit EGFR amplification also frequently present with the constitutively active EGFR variant 3 (EGFRvIII) mutant (5), which arises from the deletion of exons 2-7 of the EGFR gene, and results in a truncated, yet constitutively active, EGFR protein (6). The resultant increased downstream signaling confers enhanced glioma malignancy through multiple mechanisms, and, notably, the EGFRvIII mutation is not expressed by non-malignant tissues (6). However, although targeting EGFRvIII is a rational strategy to combat GBM, phase II clinical trials with the EGFR receptor tyrosine kinase inhibitor, erlotinib, and phase III clinical trials with the EGFRvIII vaccine, rindopepimut, have failed to robustly increase patient overall survival (OS), highlighting the immense plasticity of GBM cell populations, which diminishes treatment efficacies and hinders tumor management (5,8,9).
The coiled-coil-helix-coiled-coil-helix domain-containing protein 2 (CHCHD2), initially described as a regulator of mitochondrial respiration, has recently emerged in the contexts of non-small cell lung and renal cell carcinoma, as well as breast cancer (10-12). The CHCHD2 gene is located proximal to EGFR on chromosome 7p11.2; as such, CHCHD2 and EGFR are frequently co-amplified in non-small cell lung carcinoma (NSCLC) (10). CHCHD2 encodes a 16-kDa protein belonging to a family of nine evolutionarily conserved small mitochondrial proteins, all containing at least one CHCH domain (13,14). The CHCH domain, characterized by two CX9C motifs (two cysteines separated by nine amino acids), is necessary for simultaneous oxidative folding and protein import into the mitochondrial intermembrane space via the CHCHD4-mediated disulfide relay system (15). CHCHD2 canonically functions as a mitochondrial protein mediating cellular respiration (15-17). In addition, CHCHD2 regulates other cellular functions across a variety of biological contexts, including cell migration and the regulation of apoptosis (10,18,19). Furthermore, CHCHD2 has been implicated in mitonuclear communication through its ability to function as a nuclear transcription factor in response to hypoxia, inducing the expression of complex IV subunit 4 isoform 2 and itself maximally at 4% O2 (20). However, the subcellular localization, distribution and dynamics of CHCHD2 in GBM cells in response to hypoxia has yet not been described, at least to the best of our knowledge. Additionally, a mechanism governing its mitochondrial export and subcellular redistribution remains elusive. Thus, the functional capabilities of CHCHD2 in the context of GBM remain unexplored.
The objective of the present study was to characterize the functional capacity of CHCHD2 in GBM cells expressing EGFRvIII, as well as investigate the intracellular dynamics of CHCHD2 in response to metabolic stressors in U87 and U87vIII GBM cells. The results obtained herein indicate that subcellular distribution of CHCHD2 between mitochondria and nuclei is sensitive to the expression of EGFRvIII and hypoxia, and that CHCHD2 contributes to a number of GBM cell functions representing disease hallmarks, which may inspire therapeutic strategies targeting mitochondrial biology to potentially improve GBM tumor management.
Materials and methods
CHCHD2 gene amplification and mRNA expression analysis
The analysis of CHCHD2 gene amplification patterns across GBM tumors was performed on The Cancer Genome Atlas (TCGA) Provisional GBM database using cBioPortal (http://www.cbioportal.org) (21,22). The analysis was limited to tumor samples with available copy number alteration (CNA) data (n=577). The analysis of CHCHD2 mRNA expression levels (HG-U133A Array) was compared among GBM tumors using publicly available data via GlioVis (gliovis.bioinfo.cnio.es) (23). Tumors were stratified by variables, including tumor grade, tumor vs. non-tumor tissue, GBM subtype, glioma CpG island methylator phenotype (G-CIMP) status, isocitrate dehydrogenase 1 (IDH1) mutational status and O6-methylguanine-DNA methyl-transferase (MGMT) mutational status.
Cells and cell culture
The human U87 parental GBM cell line, as well as U87 GBM cells transduced to stably express the constitutively active EGFRvIII mutant (U87vIII) were generously provided by Dr. Nathan Price (Institute for Systems Biology, Seattle, WA, USA). The U87vIII cell line used in these studies was authenticated using STR profiling, as having a >80% identity with U-87MG ATCC (RRID:CVCL_0022), a glioblastoma of unknown origin. The U87 and U87vIII cells transfected to express the pDsRed2-Mito fluorescent mitochondrial marker (RRID:Addgene_52659) were used for a subset of immunofluorescence experiments. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 1 mM sodium pyruvate, 15 mM HEPES, non-essential amino acids, 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Media devoid of FBS were used for serum deprivation experiments. Media without phenol red were used for immunofluorescence experiments. All cell cultures were tested for mycoplasma contamination prior to orthotopic injection for experiments using mice.
Patient tissues and clinical information
GBM patient-derived cells (PDCs) were obtained from GBM xenografts established with tumor tissue from patients undergoing surgical treatment at the Mayo Clinic (Rochester, MN, USA) (24-26). These studies were approved by the Mayo Clinic Institutional Review Boards and only samples from patients who had provided prior consent for use of their tissues in research were included. A full description of the characteristics of the patients from whom the four GBM xenografts were derived and from which the PDCs were obtained and used herein, can be found in the previous study by Sarkaria et al (24).
The panel of PDCs analyzed in the present study exhibited a disparate EGFR/phosphatase and tensin homolog (PTEN) status, MGMT methylation state [methylated/unmethylated (M/U)], molecular subtype [mesenchymal/classical (M/C)], invasive characteristics in mouse orthotopic xenografts (0, low; 7, high) and sensitivity to erlotnib (0, not sensitive; 100, sensitive) (24-26). The PDCs were cultured in 3D methacrylated gelatin (GelMA) hydrogels (7% wt gelatin) without or with hyaluronic acid (HA) (6% wt gelatin, 1% wt HA) (the hydrogels were constructed in the authors' laboratory in collaboration with JWEC and BACH).
CHCHD2 protein levels in PDCs cultured in GelMA hydrogels
The total protein levels of CHCHD2 were analyzed in a panel of GBM PDCs with a disparate EGFR/PTEN status, MGMT methylation state, molecular subtype, invasive characteristics in mouse orthotopic xenografts (0, low; 7, high) and sensitivity to erlotinib (0, not sensitive; 100, sensitive) (24). The PDCs were cultured in GelMA hydrogels without HA (7% wt GelMA) or with HA (6% wt GelMA, 1% wt HA).
Hypoxic cell culture
For the hypoxic experiments, medium was pre-equilibrated overnight in a BioSpherix™ hypoxic incubator (BioSpherix, Ltd.) at designated oxygen concentrations (7, 4 and 1%) to account for the time required for oxygen-saturated media to equilibrate with the gas atmosphere (27). Standard culture conditions are designated as normoxia (20% O2). All cells were maintained at 37°C and 5% CO2.
Generation of CHCHD2 knockout (KO) cells
U87vIII CHCHD2 KO cells were derived using CRISPR-Cas9 genome engineering following published protocols (28,29). Suitable target sites within exons of the coding sequence for CHCHD2 were identified using the online WU-CRISPR design tool (30). Potential guide RNA (gRNA) oligonucleotides were obtained from Integrated DNA Technologies (IDT). Each gRNA sequence was 20 nucleotides in length and directly upstream of the protospacer adjacent motif (PAM) 5′-NGG-3′. A total of three separate gRNA expression constructs were generated by cloning phosphorylated and annealed gRNA oligos into the BbsI (New England Biolabs) site of the pSpCas9(BB)-2A-Puro expression vector (cat. no. 62988, Addgene) for the co-expression of each sgRNA with the Cas9 endonuclease. The integrity of the constructs was confirmed by plasmid sequencing (University of Illinois Urbana-Champaign Roy J. Carver Biotechnology Center).
The U87vIII cells were transfected with Cas9-CHCHD2gRNA expression constructs using Lipofectamine 2000® reagent according to the manufacturer's protocol (Thermo Fisher Scientific, Inc.). Stably transfected cells were selected with puromycin (10 μg/ml). Successful CHCHD2 protein knockout was confirmed using western blot analysis as described below. The assessment of genomic mutational status was conducted using nested PCR of the region containing the induced double-strand break using two primer sets. The PCR product was cloned into the pCR2.1-TOPO vector (K450002, Thermo Fisher Scientific, Inc.) and confirmed by sequencing (UIUC Roy J. Carver Biotechnology Center).
Measurement of oxygen consumption rate (OCR)
The mitochondrial respiration of the U87vIII CHCHD2 wild-type (WT) and KO cells was compared using the Seahorse XFp Extracellular Flux Analyzer (Agilent Technologies, Inc.). The cells were seeded at 1×104 cells/well 5 h prior to conducting the mitochondrial stress test, according to the manufacturer's protocol (Agilent Technologies). Serial applications of oligomycin (ATP synthase inhibitor, 1 μM), FCCP (protonophore, 0.5 μM), and rotenone and antimycin A (respiratory complex I and III inhibitor, respectively, 0.5 μM) over time enabled the calculation of various parameters of mitochondrial respiration in both cell lines.
Measurement of compartmentalized glutathione (GSH) redox poise
The U87vIII CHCHD2 WT and KO cells were transfected with genetically encoded, fluorescent redox biosensors targeted to the cytosol (cyto-Grx1-roGFP2) or mitochondrial matrix (mito-Grx1-roGFP2), previously described by the authors' laboratory (30). Cells expressing cytosolic or mitochondrial Grx1-roGFP2 were seeded at equal densities in standard culture medium without phenol red in μ-Slide eight-well ibiTreat microscopy chambers (ibidi GmbH). Time-lapse images were collected using a fluorescence-enabled inverted microscope (Axiovert 200 M, Carl Zeiss AG). The dual-excitation imaging of live cells used 395 and 494 nm excitation cubes, and an emission filter at 527 nm was used for both cubes. Exposure times were set to 100-200 msec, and images were obtained every 15 sec. To assess the effects of GSH synthesis inhibition on the GSH:GSH disulfide (oxidized GSH; GSSG) status, the cells were pre-treated with buthionine sulfoximine (BSO, 100 μM) (6954/100, R&D Systems, Inc.) for 24, 48, and 72 h prior to time-lapse image acquisition. Acquired images were processed using Zeiss Axiovision SE64 Rel6.8 software (AxioVision Imaging System, RRID:SCR_002677; Carl Zeiss AG), via the manual selection of three to five individual cells to obtain multiple regions of interest in each time lapse. The means of emission intensities at 527 nm were exported to Excel files and corrected by background subtraction.
Cell proliferation
The proliferation of the U87vIII CHCHD2 WT and KO cells was compared over a period of 72 h under normoxic (20% O2) and hypoxic (1% O2) culture conditions using the sulforhodamine B (SRB) assay (Abcam) according to previously published protocols (31). Briefly, the cells were seeded at equal densities in a 96-well plate, followed by fixation with 10% trichloroacetic acid (MilliporeSigma) for 1 h at 4°C at 0 and 72 h timepoints. After washing four times with water and air-drying at room temperature, 0.057% SRB solution (wt/vol in 1% acetic acid) was applied to each well and incubated for 30 min at room temperature, followed by washing four times in 1% acetic acid. After drying, bound SRB was solubilized in 10 mM Tris base solution (pH 10.5), and the plates were shaken for 30 min at room temperature. The optical density (OD) of each well was measured at 510 using a BioTek Synergy™ HT microplate reader (BioTek Instruments). OD values at 72 h were corrected by 0 h OD subtraction to account for possible variations in initial seeding densities.
Cell cytotoxicity assays
The sensitivity of the U87vIII CHCHD2 WT and KO cells to a panel of cytotoxic agents was determined using the SRB assay according to previously published protocols (31). The cytotoxicity of sulfasalazine (SSZ, xCT cystine-glutamate antiporter inhibitor; S0883, MilliporeSigma), erlotinib (Erl, EGFR receptor tyrosine kinase inhibitor; SML3621, MilliporeSigma), TMZ (DNA alkylating agent; 5.00609, MilliporeSigma) and Pac-1 (procaspase-3 activator; courtesy of Paul Hergenrother, UIUC) was assessed using doses derived from the literature (30,32).
Hydrogel preparation and measurement of cell invasion
The invasive behavior of the U87vIII CHCHD2 WT and KO cells was examined under normoxic (20% O2) and hypoxic (1% O2) culture conditions. Invasion was quantified within GelMA hydrogels using a bead invasion assay as previously described (33-35). The hydrogels used in this study were prepared using 5% wt GelMA, ~53% degree of methacrylamide functionalization determined via H1-NMR (data not shown), and photopolymerized under UV light (AccuCure LED 365 nm, 7.1 mW cm-2 for 30 sec) in the presence of a lithium acylphophinate photoinitiator. The compressive modulus of 5% wt GelMA hydrogels was measured using an Instron 5943 mechanical tester with Young's modulus obtained from the linear region of the stress-strain curve (0-10% strain) (35).
To examine invasive behavior, the U87vIII CHCHD2 WT and KO cells were seeded onto collagen-coated dextran beads (~200 μm diameter, Cytiva) at a density of 2×106 cells per 5×103 beads in 5 ml DMEM. Cell-bead suspensions were lightly shaken for 1 min, every 30 min, for 5 h to facilitate cell adhesion to the beads. The cell-coated beads were then encapsulated in pre-polymerized GelMA hydrogel solution, and bead-containing hydrogels were cultured in standard DMEM in either normoxic (20% O2) or hypoxic (1% O2) culture conditions for 7 days. The cell invasion distance was measured from the bead surface using ImageJ software (ImageJ, RRID:SCR_003070; National Institutes of Health) from images acquired using a fluorescence microscope (Leica Microsystems, Inc.) after fixing and staining cells with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) (10 μg/ml in 1X PBS). Cell invasion was reported as the mean invasion of all cells from the surface of the bead (33).
Western blot analysis
The total protein levels of CHCHD2 (1:500, cat. no. NBP1-94106, Novus Biologicals, LLC), the glutamate-cystine antiporter xCT (1:500, cat. no. ab37185, Abcam), GPx-1/2 (1:100, sc-133160, Santa Cruz Biotechnology), GPx-4 (1:100, cat. no. NBP2-75511, Novus Biologicals, LLC) and matrix metalloproteinase 2 (MMP-2; 1:500, cat. no. 10373-2-AP, Proteintech Group) were analyzed using western blot analysis. β-actin (1:1,000, cat. no. 4967, Cell Signaling Technology) was used as the loading control. Cells were lysed using standard RIPA buffer and total protein concentrations from whole cell lysates were determined using the Pierce BCA assay (Thermo Fisher Scientific, Inc.). Protein lysates were mixed 1:1 with 2X Laemmli Sample Buffer (Bio-Rad Laboratories, Inc.) (5% β-mercaptoethanol) and heated at 95°C for 5-10 min. The denatured lysates (20 μg) were loaded into 4-20% Mini-Protean® TGX™ electrophoresis gels (Bio-Rad Laboratories), and SDS-PAGE was run at 150 V for 1-1.5 h. The proteins were transferred onto a nitrocellulose membrane (Amersham; Cytiva) at 300 mA for 2 h at 4°C. The membranes were then blocked with either 5% BSA or 5% non-fat dry milk for 1 h at room temperature, and then incubated with the primary antibodies at the designated concentrations overnight at 4°C. The membranes were washed in TBS-T for 5 min three times, then incubated in HRP-linked goat anti-rabbit secondary antibody (1:2,500, cat. no. 7074, Cell Signaling Technology) at room temperature for 1.5 h. Following TBS-T washes (3×5 min each), the membranes were imaged using the SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) in an ImageQuant LAS 4010 (Cytiva). Analysis of the bands was conducted using ImageJ software v1.51 (National Institutes of Health).
Animals and GBM tumor establishment by orthotopic injection
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice [NOD scid gamma (NSG™)] mice (Jackson Laboratory) aged 8-11 weeks (20-30 g) were used in the present study with food and water at ad libitum (n=18). Consisting of an equal distribution of sexes in both groups, 9 mice were inoculated with U87vIII CHCHD2 KO and 9 with U87vIII WT. Prior to GBM cell induction, the mice were anesthetized via the intraperitoneal administration of 100 mg/kg ketamine and 10 mg/kg xylazine. The mice were distributed into groups evenly as regards sex and cell lines used, followed by injection with U87vIII WT or U87vIII CHCHD2 KO cells. The injection concentration was 1×105/μl, with the desired volume being 0.5 μl per mouse. A 0.5 μl Hamilton syringe was inserted according to the following coordinates in relation to the bregma: Rostral 0.5 mm, lateral to right 2.25 mm, and 3.3 mm lowered into brain tissue. GBM cells were infused for 30-60 sec to ensure limited injection backflow, followed by a 1-min waiting period until the removal of the needle. The incision was then closed with a small amount of 3M VetBond (Amazon.com). Post-induction, the mice were weighed daily to track percent weight change and were additionally scored for neurological symptoms of tumor formation. A weight loss of 20%, severe doming of the skull, rotational spinning and paralysis were the primary indicators for the mouse to be sacrificed. Although 16 mice were euthanized, 2 mice in the U87vIII WT group died unexpectedly overnight, possibly due to severe tumor burden (days 10 and 17 post-injection). The experiment was concluded when all mice reached the euthanasia criteria due to tumor burden symptoms (28 days post-injection). Euthanasia was administered by CO2 asphyxiation with a fill rate of 30-70% of the chamber volume per minute, verified by observable breath cessation and followed by cervical dislocation. Post-euthanasia, the brains were fixed in 4% paraformaldehyde (MilliporeSigma) for 24 h and sectioned coronally for tumor histological analysis. All animal care protocols were in accordance with the National Institutes of Health Guidelines for Care and Use of Laboratory Animals and were approved by the University of Illinois Laboratory Animal Care and Use Committee (reference no. 18058).
Statistical analysis
Differences among means were examined using the unpaired Student's t-test or one-way ANOVA, followed by post hoc Tukey's analysis where appropriate. A value of P<0.05 was considered to indicate a statistically significant difference. Variance is reported as the standard error of the mean. Odds ratios for CHCHD2 co-amplification with EGFR and log-rank tests for OS and progression-free survival (PFS) were conducted in cBioPortal (cBioPortal,RR ID:SCR_014555) (21,22). Tukey's honestly significant difference or pairwise t-tests were conducted to compare mRNA expression levels in GlioVis (23).
Results
CHCHD2 is amplified in a subset of GBM tumors and is associated with a decreased patient survival
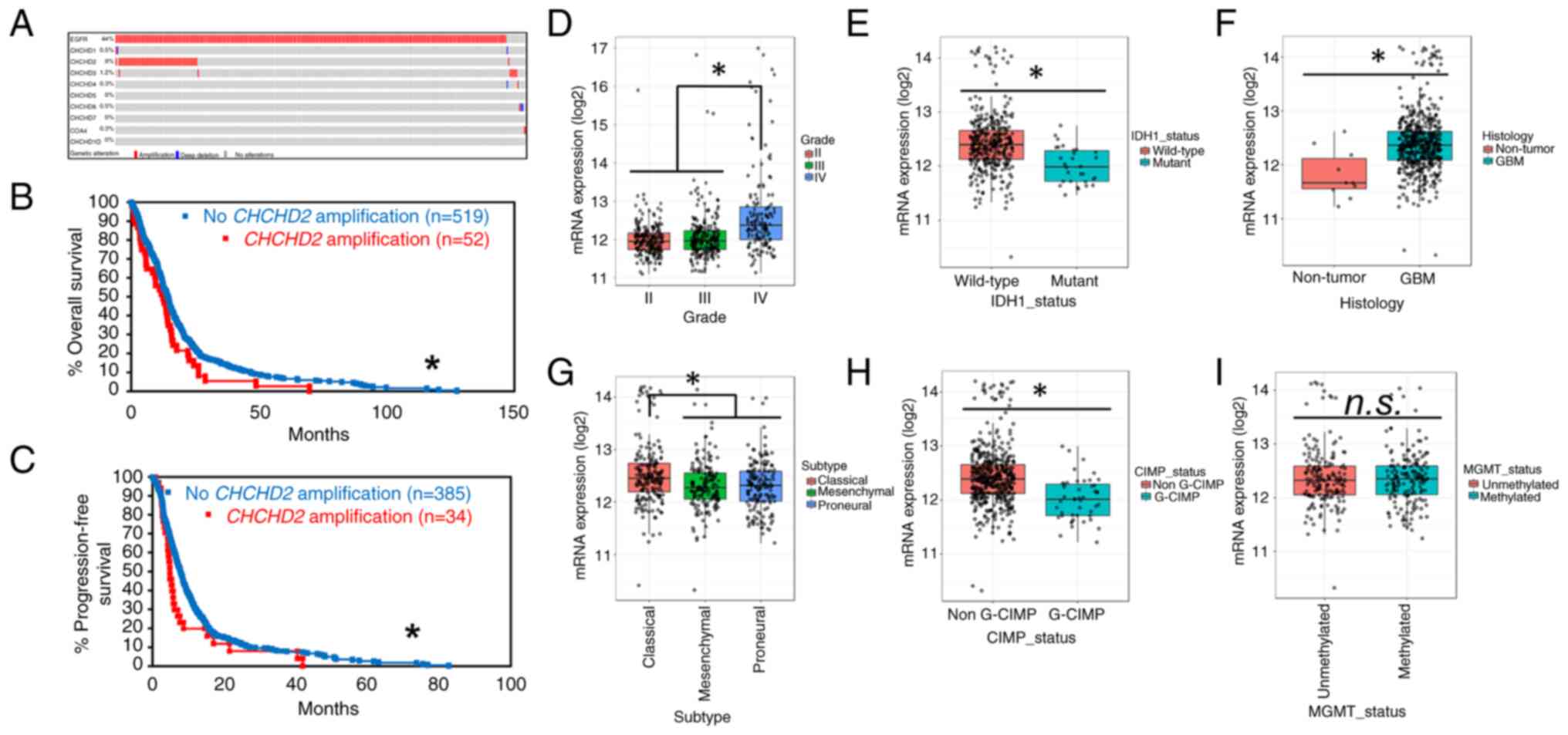
The analysis of CHCHD2 amplification patterns across GBM tumors was performed on The Cancer Genome Atlas (TCGA) Provisional GBM database using cBioPortal (http://www.cbioportal.org) (21,22). The analysis was performed on tumor samples with available CNA data (n=577). The amplification of CHCHD2 was observed in 9% of GBM tumors (Fig. 1A). Of the tumors with EGFR amplification, CHCHD2 was co-amplified in 20% of cases, with a significant tendency to co-occur (log2 odds ratio >3, P<0.001). Of the nine proteins in the CHCH domain-containing protein family, only CHCHD2 was amplified at an appreciable frequency in GBM tumors, with the next most frequently amplified CHCH protein being CHCHD3 (1.2%) (Fig. 1A). Additionally, patients with CHCHD2-amplified tumors exhibited a decreased OS (12.48 vs. 14.45 months; log-rank P=0.0223) (Fig. 1B) and PFS (4.86 vs. 7.82 months; log-rank P=0.0420) (Fig. 1C). While this effect could potentially be explained by the propensity for EGFR to co-amplify with CHCHD2, the amplification of EGFR was not solely associated with a decreased patient survival (Fig. S1). These data indicate that the amplification of the CHCHD2 gene occurs in a subset of GBM tumors, is associated with a decreased OS and PFS, and is almost always accompanied by EGFR amplification. The GlioVis data indicated that CHCHD2 mRNA expression was increased in grade IV gliomas (GBM) relative to grade II and III gliomas (Fig. 1D), as well as in IHD1-wt tumors (primary GBM) compared to IDH1-mutant (secondary GBM) (Fig. 1E). CHCHD2 expression was also increased in GBM tumors compared to non-tumor tissue (Fig. 1F). Additionally, CHCHD2 expression was increased in classical subtype tumors (Fig. 1G), likely due to the location of CHCHD2 on chromosome 7, the amplification of which defines classical GBM tumors and is accompanied by a focused predilection for EGFR amplification (36). Tumors exhibiting genome-wide promoter hypermethylation, i.e., glioma-CpG island methylator phenotype (G-CIMP), exhibited a decreased CHCHD2 mRNA expression (Fig. 1H), consistent with the epigenetic methylation-induced silencing of CHCHD2. The CHCHD2 expression levels were similar in MGMT methylated vs. unmethylated tumors (Fig. 1I).
CHCHD2 protein levels vary across GBM PDCs samples
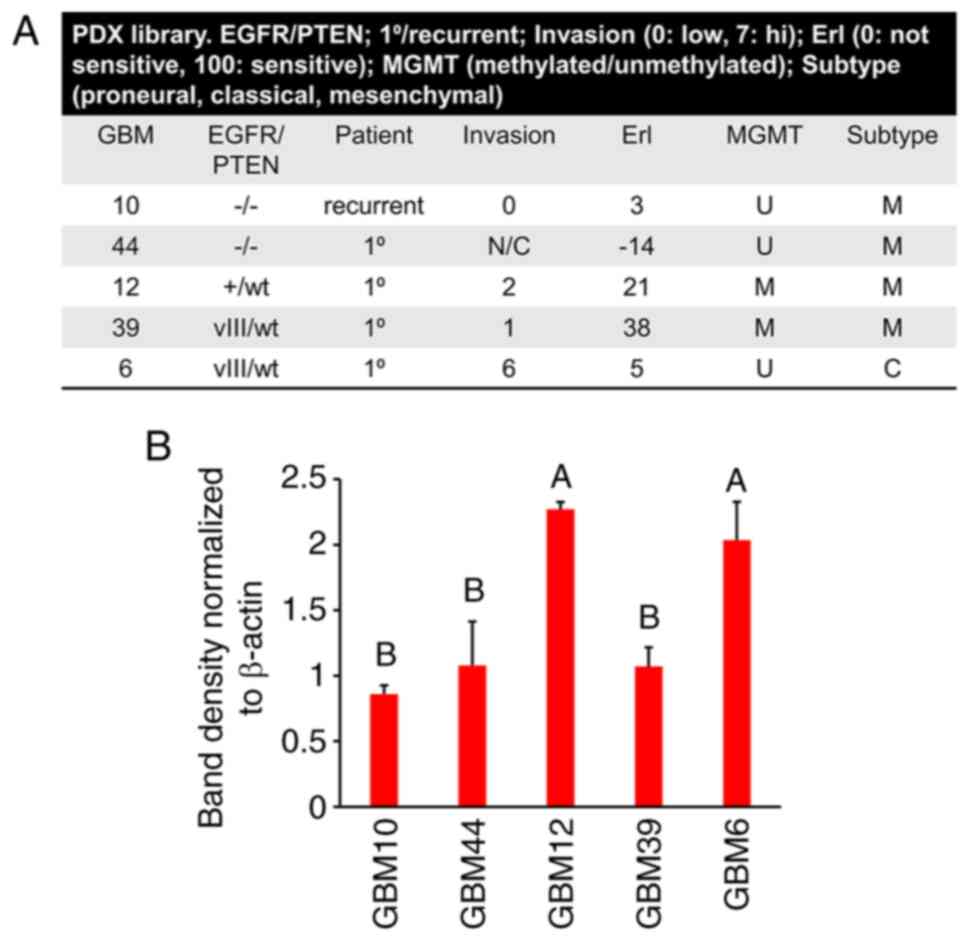
The total protein levels of CHCHD2 were analyzed in a panel of GBM PDCs cultured in GelMA hydrogels (7% wt gelatin). PDCs were characterized by a disparate EGFR/PTEN status, MGMT methylation state, molecular subtype, invasive characteristics in mouse orthotopic xenografts (0: low; 7, high) and sensitivity to erlotinib 0, not sensitive; 100, sensitive) (Fig. 2A). Included in this panel were: GBM10 (EGFR-/PTEN-), GBM44 (EGFR-/PTEN-), GBM12 (EGFR+/PTENwt), GBM39 (EGFRvIII/PTENwt) and GBM6 (EGFRvIII/PTENwt). Notably, of the PDCs analyzed in the present study, the CHCHD2 levels were greatest in GBM12 and GBM6. Notably, GBM6 expresses EGFRvIII, is the most invasive, is relatively resistant to erlotinib and is of the classical subtype (Fig. 2B). These results are in accordance with those obtained for the mRNA levels of CHCHD2, being highest in classical subtype tumors (Fig. 1G).
Knockout of CHCHD2 alters mitochondrial respiration in U87vIII cells
The gene amplification, mRNA and protein expression patterns of CHCHD2 observed across clinically relevant GBM patient samples suggested a biologically relevant role for CHCHD2 in mediating GBM cell phenotypes. To examine the essential functionality of CHCHD2 in the context of GBM, the U87vIII CHCHD2 KO cells were derived using CRISPR-Cas9 following previously published protocols (28,29) (Fig. S2), and protein knockout was validated at the protein level using western blot analysis (Fig. 3A) (28,29). CHCHD2 was first described during a computational screen as a mediator of mitochondrial respiration (16). Herein, to determine whether CHCHD2 KO affected mitochondrial respiration in GBM cells, a mitochondrial stress test was conducted on U87vIII CHCHD2 WT and KO cells using a Seahorse XFp Extracellular Flux Analyzer (Fig. 3B) (37). The CHCHD2 KO cells displayed a decreased basal and maximal OCR (Fig. 3B-D). The spare respiratory capacity was also significantly lower in the CHCHD2 KO cells (Fig. 3E), indicating a decreased ability of the U87vIII CHCHD2 KO cells to respond to the increased energy demand. Additionally, the amount of oxygen consumed coupled to ATP production by ATP synthase was significantly lower in CHCHD2 KO cells (Fig. 3F). Collectively, these data indicate that CHCHD2 is indeed required for efficient mitochondrial respiration in U87 GBM cells expressing EGFRvIII.
Knockout of CHCHD2 leads to a more reduced GSH redox pool in the mitochondrial matrix
Upon observing defects in mitochondrial respiration, it was further hypothesized that deficient electron transport chain function would result in a more oxidized intracellular redox environment manifested by increased amounts of GSSG. GSH, an enzymatically produced tripeptide of cysteine, glycine and glutamate, is the main intracellular redox buffer, which, along with thioredoxins and glutaredoxins, maintains thiol redox status (38). The GSH redox couple (reduced and oxidized glutathione, GSH and GSSG, respectively), along with glutathione peroxidase (GPx) and glutathione reductase (GR), comprises the glutathione system, which maintains thiol redox homeostasis and functions in antioxidant defense (39). The balance of reduced to oxidized glutathione (GSH:GSSG) thus represents the intracellular redox status, and can be interrogated within live cells using genetically encoded, fluorescent redox biosensors (Grx1-roGFP2) (Fig. 4A). Additionally, such probes can be targeted to various subcellular compartments, including cytosol (cyto-Grx1-roGFP2) or mitochondrial matrix (mito-Grx1-roGFP2) to measure the compartmentalized GSH redox status in live cells in real time via ratiometric fluorescence intensity measurements (Fig. 4B) (30). The percent of oxidized mito-Grx1-roGFP2 in U87vIII CHCHD2 KO cells was decreased by 15.8% compared to that measured in CHCHD2 WT cells, indicating that the pool of glutathione in the mitochondrial matrix of KO cells was more reduced than the WT counterparts (Fig. 4C). This effect was confined to the mitochondrial matrix, as the cytosolic glutathione pool was not affected by CHCHD2 KO (Fig. 4D). Treatment with BSO, an inhibitor of glutamate-cysteine ligase (the rate-limiting enzyme in GSH synthesis), led to similar oxidation of the glutathione pool over time in the mitochondrial matrix of both U87vIII CHCHD2 WT and KO cells (Fig. 4E). Additionally, the levels of components of the glutathione system, including the glutamate-cystine antiporter xCT, GPx-1/2 and GPx-4, were all unaltered in the CHCHD2 KO cells (Fig. 4F). These data demonstrate a role for CHCHD2 in mediating the GSH redox balance specifically in the mitochondrial matrix, albeit through a mechanism not involving GSH synthesis, cystine import through xCT, or reduced GSH flux through glutathione peroxidases 1, 2 or 4.
U87vIII cell growth and invasion are negatively affected by CHCHD2 KO under both normoxic and hypoxic conditions
The observed deficiencies in mitochondrial respiration led to the hypothesis that U87vIII GBM cell growth would be negatively affected by CHCHD2 KO. To examine this hypothesis, the U87vIII CHCHD2 WT and KO cells were incubated in either standard oxygen culture conditions (normoxia, 20% O2) or pathophysiologically relevant hypoxia (1% O2). Utilizing the SRB colorimetric assay (31) to assess cell growth over a period of 72 h, CHCHD2 WT cell growth was observed to be significantly increased under hypoxic conditions (Fig. 5A). CHCHD2 KO cell growth was decreased compared to the normoxic control (Fig. 5A). Additionally, the growth-inducing effect of hypoxia on U87vIII cells was abrogated upon CHCHD2 KO (Fig. 5A). These results demonstrate that CHCHD2 is involved not only in mediating U87vIII cell growth under normoxic conditions, but also plays a role in the initial increased cell proliferation observed in hypoxic cells.
The rapid, diffuse invasion of GBM cells into tumor margins and into the surrounding brain parenchyma represents a major obstacle impeding effective tumor treatment. Herein, to determine whether CHCHD2 knockout affects GBM cell invasion, a bead invasion assay in GelMA hydrogels (Young's modulus 2.9±0.45 kPa) was used as a three-dimensional culture platform to model biophysical aspects of the brain parenchyma and GBM cell invasion (33,34). This technique enables the monitoring of cell invasion from a defined starting point in a spatiotemporal manner in normal as well as hypoxic three-dimensional culture conditions (Fig. 5B). Hypoxia (1% O2) stimulated U87vIII cell invasion over long-term culture (7 days; Fig. 5C). In contrast, the U87vIII CHCHD2 KO cells displayed minimal cell invasion under both normoxic and hypoxic conditions, with the hypoxia-induced invasion observed in CHCHD2 WT cells being abrogated (Fig. 5C). Furthermore, the CHCHD2 KO cells displayed decreased basal levels of pro-MMP-2 (Fig. 5D), a key protein involved in the breakdown of extracellular matrix. These data demonstrate a role for CHCHD2 in mediating U87vIII cell invasion potentially involving MMP-2, particularly in response to hypoxia.
CHCHD2 knockout enhances U87vIII sensitivity to a variety of cytotoxic drugs
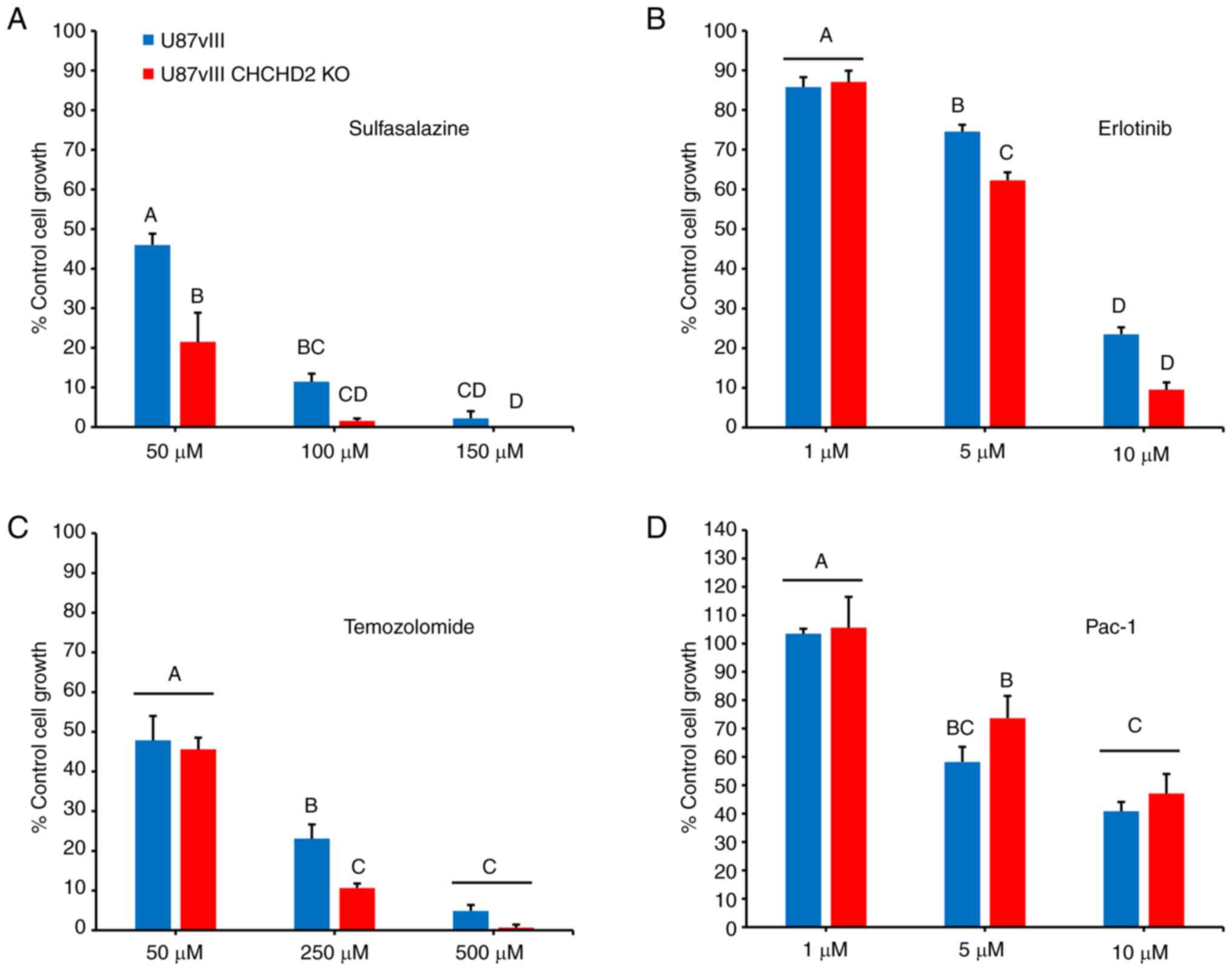
To determine the effects of CHCHD2 KO on cellular resistance to various chemotherapeutic agents, the cells were treated with increasing concentrations of a panel of drugs, and cytotoxicity was assessed using the SRB assay (31). Included in this panel were: TMZ, a DNA-alkylating agent and the standard-of-care chemotherapy administered to patients with GBM (38); Erl, a receptor tyrosine kinase inhibitor that inhibits EGFR and EGFRvIII tyrosine kinase activity (8); SSZ, an inhibitor of the cell membrane xCT antiporter, which couples the export of the amino acid glutamate with the import of cystine, thus depriving cells of the rate-limiting substrate to synthesize reduced GSH (40-42); and Pac-1, a novel activator of apoptosis which acts on procaspase-3 (32). The results (Fig. 6) demonstrated that the CHCHD2 KO cells were more susceptible to treatment with TMZ, Erl, and most significantly, SSZ. However, CHCHD2 KO had no effect on cellular sensitivity to treatment with Pac-1 (Fig. 6D), consistent with the findings of a previous study, demonstrating that CHCHD2 regulates apoptosis upstream of procaspase-3 activity in the apoptotic cascade (19). These results demonstrate a role for CHCHD2 in mediating cell sensitivity to various drugs relevant to GBM treatment, highlighting CHCHD2 as a promising avenue for future investigations.
CHCHD2 knockout in GBM tumors affects the overall survival of mice
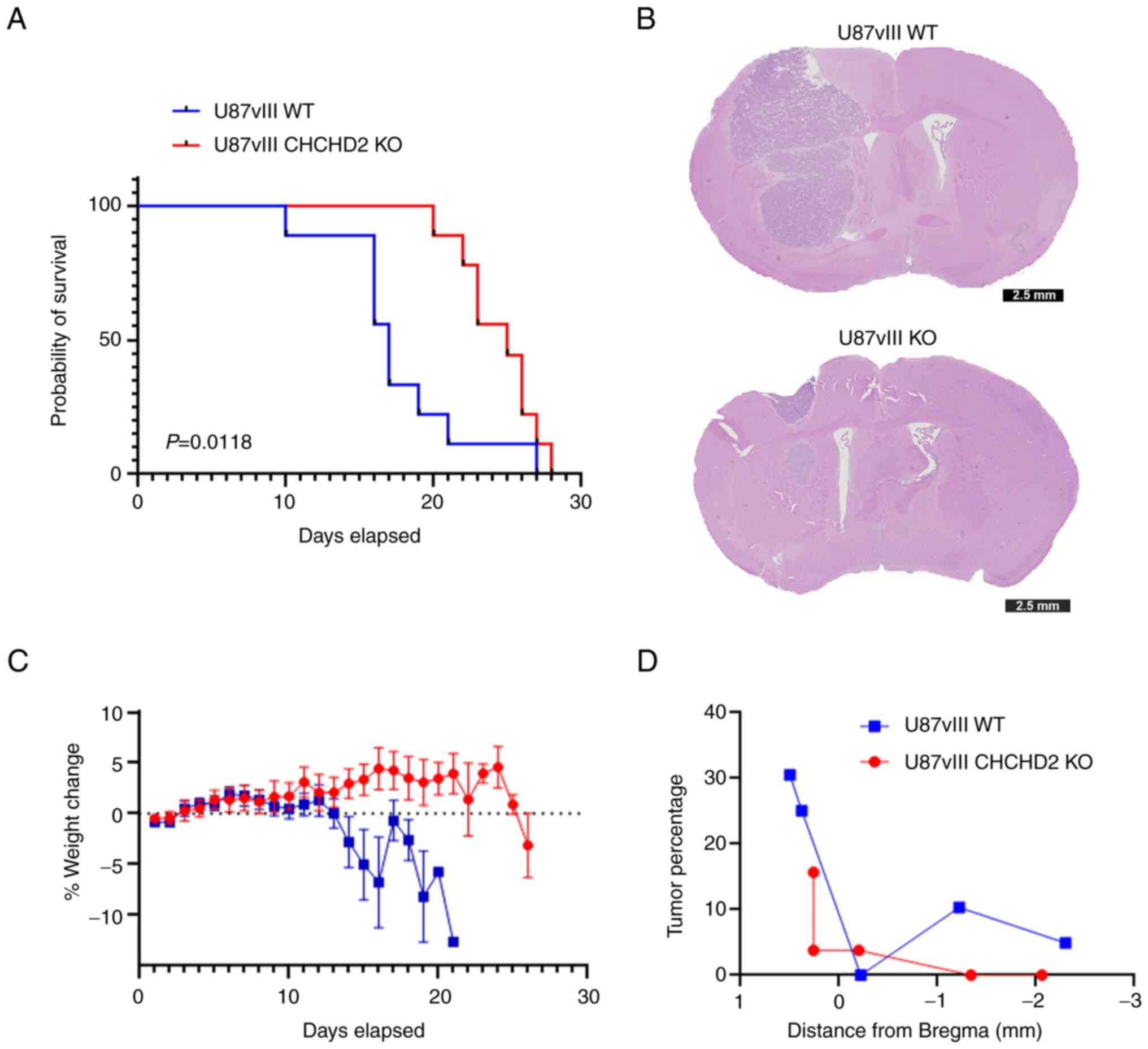
To determine the effects of CHCHD2 on tumor growth in vivo, NSG mice were orthotopically injected with U87vIII WT and CHCHD2 KO cells. Mice bearing U87vIII WT tumors presented with a median survival of 17 days vs. 25 days for U87vIII CHCHD2 KO tumor-bearing mice (log-rank P=0.0118) (Fig. 7A). Representative coronal sections of mice sacrificed on day 16 post-injection demonstrated substantial tumor growth and infiltration into the surrounding brain (Fig. 7B). The mice injected with U87vIII WT cells exhibited a rapidly diminishing health by day 13 of the study, as evidenced by a decrease in weight (Fig. 7C). Furthermore, the U87vIII WT cells consistently produced tumors occupying a larger percentage of parenchyma compared to the CHCHD2 KO-derived tumors (Fig. 7D). Overall, these data indicate a role for CHCHD2 in promoting the progression of GBM tumors in mice.
Discussion
Mitochondria, in addition to regulating cellular energy conservation, serve as signaling organelles. The vast majority of mitochondrial proteins are encoded by nuclear genes, necessitating the ability for mitochondria to communicate their status to the nucleus in response to metabolic perturbations. Disturbances in ATP and ROS production, damage to mitochondrial DNA, and aberrations in mitochondrial protein folding induce a mitonuclear retrograde signaling pathway by which mitochondria communicate with the nucleus to induce changes in nuclear gene expression in order to maintain metabolic homeostasis (43). Several proteins have been demonstrated to participate in inter-organelle signaling between mitochondria and the nucleus, including p53, fumarase, the pyruvate dehydrogenase complex and CHCHD2 (15,44-46). CHCHD2 presented as a promising target to investigate, for the following reasons: i) Its proximity to and frequency of co-amplification with EGFR with NSCLC and GBM (10), both of which have been characterized as relatively oxidative vs. fermentative tumors (47); ii) its mRNA expression patterns across glioma grade and tumor vs. non-tumor tissue; iii) its reported oxygen-sensitive transcription factor activity (15,20); and iv) its pleiotropic roles mediating cellular functions reminiscent of cancer hallmarks, including proliferation, migration and invasion, and the inhibition of apoptosis (15,18,19). The present study demonstrated that CHCHD2 was involved in mediating therapeutic sensitivity, as well as cell growth and invasion in in vitro and in vivo models of GBM.
The knockout of CHCHD2 in U87vIII GBM cells resulted in decreased baseline respiration as well as spare respiratory capacity, an effect corroborated by multiple studies which have characterized CHCHD2 as a canonical regulator of mitochondrial respiration (10,15-17). While metabolic reprogramming towards increased glycolytic flux to favor increased cell proliferation is a recognized hallmark of cancer, functional mitochondria remain essential in maintaining malignant cell bioenergetics in particular tumors, including those of lung and brain (47-49). Notably, mitochondrial spare respiratory capacity has been positively associated with glioma stem cell resistance to radiotherapy (50). Thus, the decreased spare respiratory capacity measured in U87vIII CHCHD2 KO cells may partially contribute to their increased sensitivity to treatment with TMZ, Erl, and SSZ.
Additionally, CHCHD2 has ben demonstrated to mediate mitochondrial outer membrane permeabilization (MOMP) (19), the 'point of no return' during the intrinsic pathway of apoptosis. Indeed, in the present study, the U87vIII CHCHD2 KO cells exhibited an enhanced sensitivity to TMZ, Erl and SSZ. The observation that no increase in cytotoxicity was found in the CHCHD2 KO cells treated with Pac-1, an activator of procaspase-3, which acts downstream of MOMP in the apoptotic cascade (32), further corroborates the demonstrated role of CHCHD2 early in the regulation of apoptosis.
Notably, as evidenced by the use of mito-Grx1-roGFP2, the mitochondrial GSH pool was more reduced in response to CHCHD2 KO compared to WT cells, an effect independent of GSH biosynthesis. It should be noted that roGFP2 itself is not directly oxidized by reactive oxygen species (ROS), but rather equilibrates with the local GSH redox potential, which is influenced by the GSH:GSSG ratio, and in turn is influenced by activity of various ROS-scavenging enzymes that use GSH as a cofactor. As previously demonstrated, in 293/293T cells, CHCHD2 knockdown was accompanied by the decreased expression of superoxide dismutase 2 and the loss of glutathione peroxidase expression (15). However, herein, no changes were observed in the protein levels of GPx-1/2, Gpx-4, nor the glutamate-cystine antiporter xCT upon CHCHD2 knockdown in U87vIII cells.
Incubation under hypoxic conditions increased U87vIII CHCHD2 WT cell proliferation over a period of 3 days, and increased invasion over a period of 7 days. Using the GelMA hydrogel platforms described herein, a previous study demonstrated that hypoxic U87vIII cells exhibited an increased cell proliferation until day 5, at which point cell proliferation stalled, while invasion continued to increase up to day 7 (35). Notably, CHCHD2 KO abrogated the increased cell growth and invasion in hypoxia exhibited by WT cells. The observed decrease in U87vIII CHCHD2 KO cell growth is not likely due to an increase in apoptosis, as evidence has indicated that shRNA-mediated CHCHD2 knockdown does not alter the cellular levels of poly (ADP-ribose) polymerase (PARP) (15). Deficiencies in mitochondrial respiration and ATP production are likely responsible for hampering cell proliferative capacity, and may also be implicated in the suppression of cell invasion, as migration and invasion throughout parenchyma is an energy-expensive process, particularly when moving through dense extracellular matrices (51). The observed decrease in pro-MMP-2 expression in CHCHD2 KO cells provides further explanation for their decrease in invasive capacity. Another study highlighted CHCHD2 as an activator of NIH3T3 fibroblast migration via the AKT-RhoA/ROCK-JNK cascade (18). Critically, the effects of CHCHD2 observed in vitro were closely mirrored in vivo, as mice orthotopically injected with U87vIII CHCHD2 KO cells presented with a decreased tumor burden and an improved survival time.
To the best of our knowledge, the present study is the first to describe the functional relevance of CHCHD2 in GBM. The data obtained indicate that CHCHD2 is essential for mitochondrial respiration and maintenance of the mitochondrial GSH status in U87vIII cells. Additionally, these findings demonstrate a critical role for CHCHD2 in mediating cell growth and invasion under normoxic and hypoxic conditions, and resistance to various cytotoxic agents, underscoring CHCHD2 as a mediator of the GBM malignant phenotype. The functional outcomes investigated herein relate primarily to the mitochondrial functions of CHCHD2. As a protein implicated in mitonuclear communication in response to hypoxia, the nuclear functions of CHCHD2 in GBM cells and the panel of nuclear genes it regulates are of equal importance and comprise topics for future research. The nuclear function of CHCHD2 has been previously demonstrated to function in concert with other transcription factors, namely recombining binding protein suppressor of hairless (RBPJ) (20), the main downstream effector protein of the Notch signaling pathway, which itself has been implicated in the maintenance of glioma stem cell maintenance and viability (52). Future studies are required to identify the complement of genes regulated by CHCHD2 in both normal oxygen conditions, as well as under hypoxic conditions.
Limitations of the present study include the use of a single cell line to determine the effects of knocking out the expression of CHCHD2 via CRISPR-Cas9 genome engineering. Consistent with GBM tumors, the PDCs include multiple cell types, thereby precluding the knockout of CHCHD2 in the heterogeneous patient-derived samples. Regardless, the combined cell line and patient-derived data clearly indicate that a more in-depth understanding of both the nuclear and mitochondrial functions of CHCHD2 may identify the mechanisms through which this intriguing protein involved in mitonuclear retrograde signaling may be manipulated to improve the outcomes of patients with GBM.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JCL, BACH, AJS and HRG were involved in the conception and design of the study. JCL, VLK, JWEC and JNS were involved in the development of the study methodology. JCL, PLH, VLK, JEC, JS, AR, MS, AJS and HRG were involved in the acquisition of data (provided animals, provided facilities and performed experiments). JCL, PLH, VLK, MS, BACH, AJS and HRG were involved in the analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis). JCL, PLH, BACH, AJS and HRG were involved in the writing, reviewing and/or revision of the manuscript. HRG supervised the study. JCL and HRG confirm the authenticity of the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The use of human tumor tissues was were approved by the Mayo Clinic Institutional Review Boards and only samples from patients who had provided prior consent for use of their tissues in research were included. All animal care protocols were in accordance with National Institutes of Health Guidelines for Care and Use of Laboratory Animals and were approved by the University of Illinois Laboratory Animal Care and Use Committee (reference no. 18058).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank Professor Romana Nowak (University of Illinois Urbana-Champaign) for the use of the BioSpherixTM hypoxic incubator and Professor Zeynep Madak-Erdogan (University of Illinois Urbana-Champaign) for the use of the Seahorse XFp Extracellular Flux Analyzer.
Funding
The present study was partially supported by the National Cancer Institute of the National Institutes of Health (grant nos. R01 CA256481 and R01 CA197488). The authors also acknowledge additional funding provided by the Department of Chemical and Biomolecular Engineering, as well as the Cancer Center at Illinois at the University of Illinois Urbana-Champaign.
References
|
Barnholtz-Sloan JS, Ostrom QT and Cote D: Epidemiology of brain tumors. Neurol Clin. 36:395–419. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, et al: Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 318:2306–2316. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Omuro A and DeAngelis LM: Glioblastoma and other malignant gliomas: A clinical review. JAMA. 310:1842–1850. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al: Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 164:550–563. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, Ashby L, Mechtler L, Goldlust SA, Iwamoto F, et al: Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 18:1373–1385. 2017. View Article : Google Scholar | |
|
Gan HK, Kaye AH and Luwor RB: The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 16:748–754. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Parker JJ, Canoll P, Niswander L, Kleinschmidt-DeMasters BK, Foshay K and Waziri A: Intratumoral heterogeneity of endogenous tumor cell invasive behavior in human glioblastoma. Sci Rep. 8:180022018. View Article : Google Scholar : | |
|
Raizer JJ, Abrey LE, Lassman AB, Chang SM, Lamborn KR, Kuhn JG, Yung WK, Gilbert MR, Aldape KA, Wen PY, et al: A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 12:95–103. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Raizer JJ, Giglio P, Hu J, Groves M, Merrell R, Conrad C, Phuphanich S, Puduvalli VK, Loghin M, Paleologos N, et al: A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J Neurooncol. 126:185–192. 2016. View Article : Google Scholar : | |
|
Wei Y, Vellanki R, Coyaud É, Ignatchenko V, Li L, Krieger J, Taylor P, Tong J, Pham NA, Liu G, et al: CHCHD2 is coamplified with EGFR in NSCLC and regulates mitochondrial function and cell migration. Mol Cancer Res. 13:1119–1129. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng Q, Qu D, Lu Z and Zhang L: Knockdown of CHCHD2 inhibits migration and angiogenesis of human renal cell carcinoma: A potential molecular marker for treatment of RCC. Oncol Lett. 17:765–772. 2019.PubMed/NCBI | |
|
Aras S, Maroun MC, Song Y, Bandyopadhyay S, Stark A, Yang Z, Long MP, Grossman LI and Fernández-Madrid F: Mitochondrial autoimmunity and MNRR1 in breast carcinogenesis. BMC Cancer. 19:4112019. View Article : Google Scholar : PubMed/NCBI | |
|
Modjtahedi N, Tokatlidis K, Dessen P and Kroemer G: Mitochondrial proteins containing Coiled-Coil-Helix-Coiled-Co il-Helix (CHCH) domains in health and disease. Trends Biochem Sci. 41:245–260. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou ZD, Saw WT and Tan EK: Mitochondrial CHCHD-containing proteins: Physiologic functions and link with neurodegenerative diseases. Mol Neurobiol. 5:5547–5549. 2017. View Article : Google Scholar | |
|
Aras S, Bai M, Lee I, Springett R, Hüttemann M and Grossman LI: MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion. 20:43–51. 2015. View Article : Google Scholar | |
|
Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z and Mootha VK: A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 5:e10005902009. View Article : Google Scholar : | |
|
Meng H, Yamashita C, Shuba-Fukushima K, Inoshita T, Funayama M, Sato S, Hatta T, Natsume T, Umitsu M, Takagi J, et al: Loss of Parkinson's disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat Commun. 8:155002017. View Article : Google Scholar : PubMed/NCBI | |
|
Seo M, Lee WH and Suk K: Identification of novel cell migration-promoting genes by a functional genetic screen. FASEB J. 24:464–478. 2010. View Article : Google Scholar | |
|
Liu Y, Clegg HV, Leslie PL, Di J, Tollini LA, He Y, Kim TH, Jin A, Graves LM, Zheng J and Zhang Y: CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ. 22:1035–1046. 2015. View Article : Google Scholar : | |
|
Aras S, Pak O, Sommer N, Finley R Jr, Hüttemann M, Weissmann N and Grossman LI: Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 41:2255–2266. 2013. View Article : Google Scholar | |
|
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al: The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 2012. View Article : Google Scholar | |
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al: Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6:p112013. View Article : Google Scholar | |
|
Bowman RL, Wang Q, Carro A, Verhaak RGW and Squatrito M: GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 19:139–141. 2017. View Article : Google Scholar | |
|
Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A and James CD: Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 12:2264–2271. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, Galanis E, Giannini C, Wu W, Dinca EB and James CD: Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 6:1167–1674. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Vaubel RA, Tian S, Remonde D, Schroeder MA, Mladek AC, Kitange GJ, Caron A, Kollmeyer TM, Grove R, Peng S, et al: Genomic and phenotypic characterization of a broad panel of patient-derived xenografts reflects the diversity of glioblastoma. Clin Cancer Res. 26:1094–1104. 2020. View Article : Google Scholar | |
|
Hüttemann M, Lee I, Liu J and Grossman LI and Grossman LI: Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 274:5737–5748. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA and Zhang F: Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8:2281–2308. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Wong N, Liu W and Wang X: WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16:2182015. View Article : Google Scholar : PubMed/NCBI | |
|
Kolossov VL, Beaudoin JN, Ponnuraj N, DiLiberto S, Hanafin WP, Kenis PJA and Gaskins HR: Thiol-based antioxidants elicit mitochondrial oxidation via respiratory complex III. Am J Physiol Cell Physiol. 309:C81–C91. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Vichai V and Kirtikara K: Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 1:1112–1116. 2006. View Article : Google Scholar | |
|
Joshi AD, Botham RC, Schlein LJ, Roth HS, Mangraviti A, Borodovsky A, Tyler B, Joslyn S, Looper JS, Podell M, et al: Synergistic and targeted therapy with a procaspase-3 activator and temozolomide extends survival in glioma rodent models and is feasible for the treatment of canine malignant glioma patients. Oncotarget. 8:80124–80138. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Chen JE, Lumibao J, Blazek A, Gaskins HR and Harley B: Hypoxia activates enhanced invasive potential and endogenous hyaluronic acid production by glioblastoma cells. Biomater Sci. 6:854–862. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Chen JE, Pedrons S and Harley BAC: The combined influence of hydrogel stiffness and matrix-bound hyaluronic acid content on glioblastoma invasion. Macromol Biosci. 17: View Article : Google Scholar : 2017. | |
|
Chen JE, Pedron S, Shyu P, Hu Y, Sarkaria JN and Harley BAC: Influence of hyaluronic acid transitions in tumor microenvironment on glioblastoma malignancy and invasive hehavior. Front Mater. 5:392018. View Article : Google Scholar | |
|
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al: The somatic genomic landscape of glioblastoma. Cell. 155:462–477. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN and Jastroch M: Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 547:309–354. 2014. View Article : Google Scholar | |
|
Hanschmann EM, Godoy JR, Berndt C, Hudemann C and Lillig CH: Thioredoxins, glutaredoxins, and peroxiredoxins-molecular mechanisms and health significance: From cofactors to antioxidants to redox signaling. Antioxid Redox Signal. 19:1539–1605. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Estrela JM, Ortega A and Obrador E: Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 43:143–181. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Chen L, Li X, Liu L, Yu B, Xue Y and Liu Y: Erastin sensitizes glioblastoma cells to temozolomide by restraining xCT and cystathionine-γ-lyase function. Oncol Rep. 33:1465–1474. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Huberfeld G and Vecht CJ: Seizures and gliomas-towards a single therapeutic approach. Nat Rev Neurol. 12:204–216. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Eisenberg-Bord M and Schuldiner M: Ground control to major TOM: Mitochondria-nucleus communication. FEBS J. 284:196–210. 2017. View Article : Google Scholar | |
|
Yogev O, Naamati A and Pines O: Fumarase: A paradigm of dual targeting and dual localized functions. FEBS J. 278:4230–4242. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Zhuang J, Wang PY, Huang X, Chen X, Kang JG and Hwang PM: Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc Natl Acad Sci USA. 110:17356–17361. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E and Michelakis ED: A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 158:84–97. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Courtney KD, Bezwada D, Mashimo T, Pichumani K, Vemireddy V, Funk AM, Wimberly J, McNeil SS, Kapur P, Lotan Y, et al: Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28:793–800.e2. 2018. View Article : Google Scholar : | |
|
Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XI, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al: Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15:827–237. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Pavlova NN and Thompson CB: The emerging hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, Popescu R, Della Donna L, Evers P, Dekmezian C, et al: Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 108:16062–16067. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Zanotelli MR, Goldblatt ZE, Miller JP, Bordeleau F, Li J, VanderBurgh JA, Lampi MC, King MR and Reinhart-King CA: Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol Biol Cell. 29:1–9. 2018. View Article : Google Scholar : | |
|
Xie Q, Wu Q, Kim L, Miller TE, Liau BB, Mack SC, Yang K, Factor DC, Fang X, Huang Z, et al: RBPJ maintains brain tumor-initiating cells through CDK9-mediated transcriptional elongation. J Clin Invest. 126:2757–2772. 2016. View Article : Google Scholar : |