Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis
- Authors:
- Published online on: December 28, 2012 https://doi.org/10.3892/mmr.2012.1254
- Pages: 761-766
Abstract
Introduction
Oral squamous cell carcinoma (OSCC) represents ~3% of all malignancies and accounts for >274,000 newly diagnosed cancer cases every year worldwide (1). Notably, this number has not changed significantly for >50 years (2). Therefore, an early detection method for OSCC is needed to increase long-term patient survival.
Non-coding RNAs (ncRNAs) are emerging new regulators of diverse biological functions, playing an important role in oncogenesis and tumor progression. NcRNAs may be grouped into two major classes based on their transcript sizes: small ncRNAs (<200 bp) and long ncRNAs (lncRNAs) (≥200 bp) (3,4). Small ncRNAs comprise a broad range of RNA species, many associated with 5′ or 3′ regions of genes, including the well-documented microRNAs (miRNAs) that are detectable in the blood, sputum and urine of cancer patients (5–7).
lncRNAs have been found to be dysregulated in a wide range of human diseases and disorders, including various types of cancer, such as breast (8), colorectal (9), prostate cancer (10), hepatocellular carcinoma (11,12), leukemia (13) and melanoma (14). Hence, lncRNAs may also be associated with oral cancer.
There have been several studies on the aberrant expression of miRNAs in OSCC (15,16), which have been suggested to be potential biomarkers for disease diagnosis (17,18). However, there is no information regarding the expression of lncRNAs in OSCC. In this study, we analyzed the expression profiles of matched samples of OSCC and nonmalignant tissue using a real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) in cancer-related lncRNAs and identified an expression signature of selected lncRNAs that were predominantly upregulated in the most OSCC samples. Saliva, a local biofluid for oral cancer, has been shown to harbor clinical discriminatory proteomic and transcriptomic biomarkers. We tested the hypothesis that lncRNAs are secreted in saliva and may be used clinically to detect oral cancer. lncRNAs provide novel insights into the general molecular mechanisms associated with malignant transformations in oral tissue and indicate potential new targets for the diagnosis and treatment of OSCC.
Altered expression of lncRNAs has been documented in different types of human cancer (19–21) prompting an increasing interest in their use as biomarkers for diagnosis and prognosis as well as potential therapeutic targets (22,23).
No information regarding the expression of lncRNAs in OSCC has been previously published. Hence, we identified six well-documented lncRNAs associated with cancer from previous studies, i.e., MALAT-1 (8,24), HOTAIR (8), NEAT-1 (25), HULC (26), MEG-3 (27) and UCA1 (28), as samples to evaluate their expression in tissues and saliva from OSCC patients.
Materials and methods
Tissue samples
A total of 20 tumor tissue samples were obtained from OSCC patients who underwent radical tumor resection at the Department of Oral and Maxicallifacial Surgery, the Affiliated Stomatology Hospital of Sun Yat-sen University (Guangzhou, China) and were used in the experiments. The study was approved by the ethics committee of the Institute of Stomatological Research at the university. Of the 20 tumor tissue samples, 8 matched samples of tumor and adjacent nonmalignant tissue were obtained from 4 OSCC patients, as well as 12 samples with/without metastasis. Saliva samples (1 ml) were obtained from each patient prior to surgery and mixed with 3 ml RNAlater (Ambion, Foster City, CA, USA). Fresh tissue samples were obtained from the patients, with informed consent, and cut into fragments <0.5 cm in any single dimension. The tissues were then immersed into 2 ml RNAlater. All the saliva and tissue samples were frozen 30 min following surgery and stored in liquid nitrogen until use. Tissue sections from each OSCC sample were reviewed and classified by a pathologist. Tumor tissue fragments subsequently processed for RNA isolation should contain ≥80% neoplastic cells, as determined by hematoxylin and eosin (H&E) staining of tissue sections and the adjacent nonmalignant tissues were also validated as no neoplastic cells were present. The clinical status of the disease was heterogeneous across the patients: seven patients showed metastasis, three showed locally advanced disease and six showed localized tumors. The anatomopathological data of the OSCC patients are listed in Table I.
RT-qPCR of six lncRNAs in OSCC tissue and saliva samples
The OSCC tissue and saliva samples were thawed from the liquid nitrogen before use. The tissues were retrieved from RNAlater with sterile forceps. Excess RNAlater solution was blotted away with an absorbent lab wipe. Saliva samples were centrifuged at 20,000 × g for 5 min at 4°C and the supernatants were discarded. Total RNA from tissue and saliva samples were extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Reverse transcription reactions were performed using 1 μg total RNA with the PrimeScript® RT reagent kit (Takara Bio, Shiga, Japan) using random hexamer primers. Real-time PCR was then performed using the SYBR® Premix Dimer Eraser kit (Takara Bio). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was evaluated as a housekeeper gene for the qPCR reaction using the commercial PCR primers (Takara Bio). Each test was performed in triplicate and the 2−ΔCt method was used to calculate the expression of each lncRNA in all the tissue and saliva samples. All the reactions were performed on a Bio-Rad CFX-96 real-time PCR system (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. The experiments were in strict compliance with the Minimum Information about Quantitative Real-Time PCR Experiments (MIQE) guidelines. The nucleotide sequence of the qPCR primers are listed in Table II.
Statistical analysis and algorithm of figure generation
The difference in tissue lncRNA expression among the patients was analyzed using the Mann-Whitney U test concerning clinical parameters such as median age, gender and lymph node metastasis. For paracarcinoma-carcinoma paired tissues, the difference in lncRNA expression was evaluated with a Wilcoxon matched-pairs signed-ranks test. All the statistical analyses were performed with STATA 10.0 (StataCorp LP, College Station, TX, USA). P<0.05 was considered to indicate a statistically significant difference. The lncRNA expression heat-maps shown in Figs. 1A and 2A were generated by matrix2png (http://chibi.ubc.ca/matrix2png/bin//matrix2png.cgi). Rows of each gene expression matrix were normalized to have mean zero, variance one.
Results
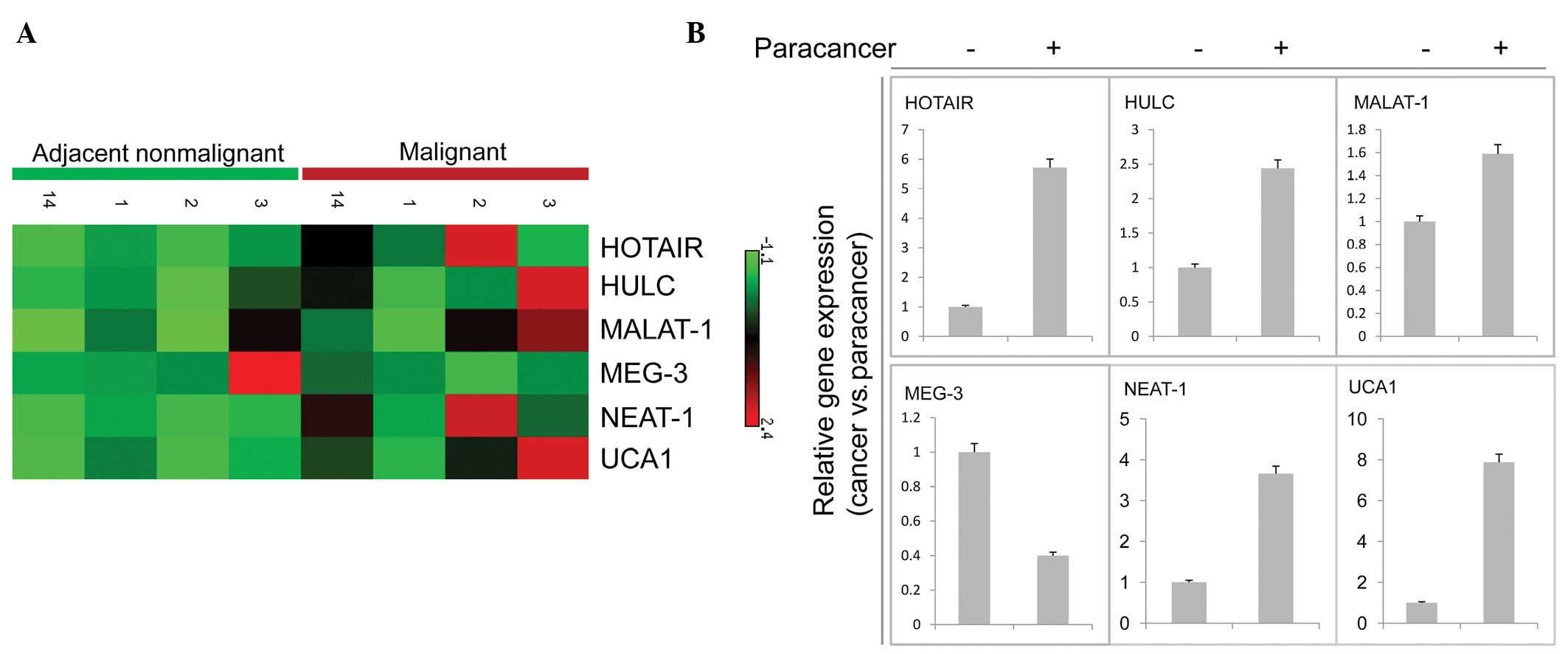
lncRNA expression in cancer and matched adjacent non-tumor tissue samples
We firstly compared the expression profiles from four OSCC samples with those from their matched adjacent non-tumor tissue samples. We found that most of the selected transcripts (4/6) were upregulated in tumors relative to matched adjacent nonmalignant tissue (Fig. 1A and B). One gene, MEG-3, was downregulated in cancer compared with its adjacent nonmalignant tissue (Fig. 1A and B). However, we analyzed a gene (MALAT-1) identified by Ji et al(24), which did not have a significantly different expression between primary tumors and matched adjacent nonmalignant tissues (Fig. 1B). An increased expression of the lncRNA MALAT-1 was firstly observed in metastatic non-small cell lung cancer (24), followed by endometrial stromal sarcoma of the uterus (29) and in six additional types of cancer, including hepatocellular carcinoma, breast, pancreas, lung, colon and prostate cancer (30).
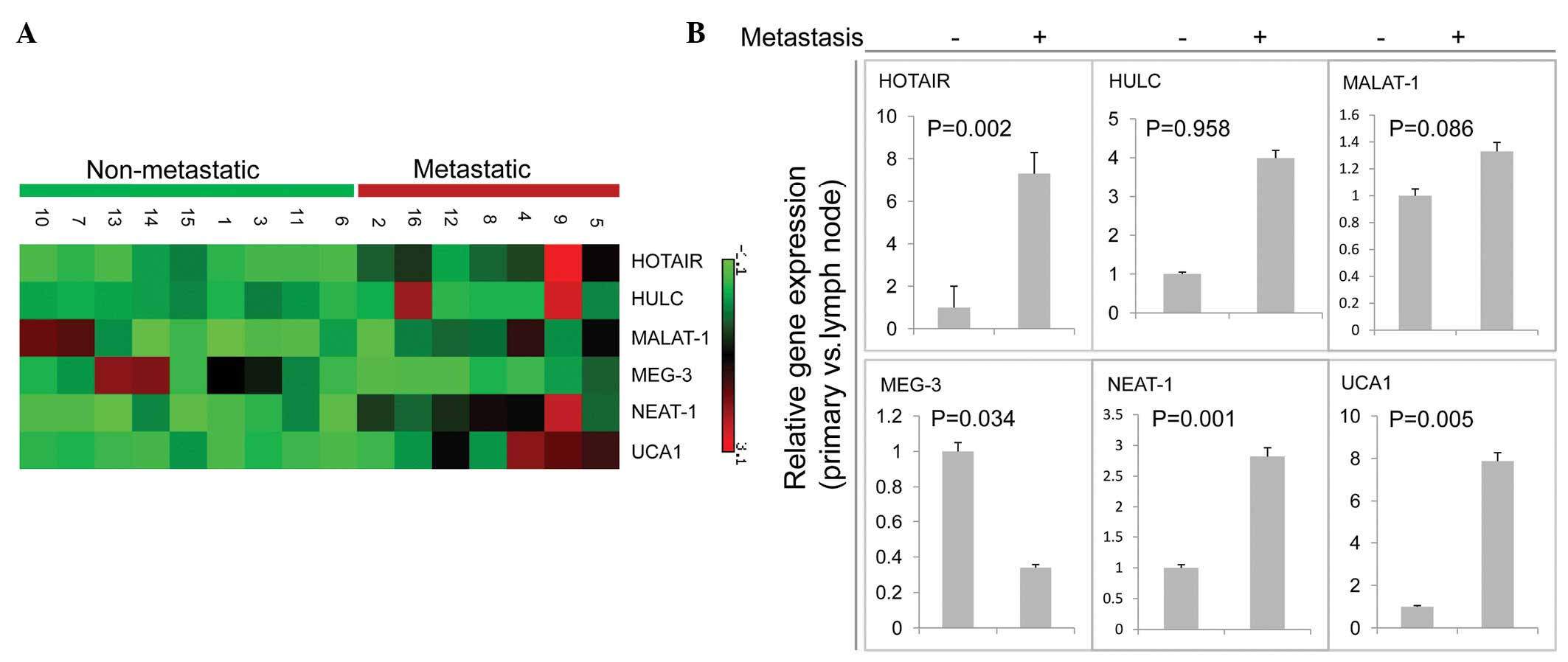
lncRNAs expression in nonmetastatic and metastatic cancer tissue samples
Next, we focused on the expression of these genes in metastatic tumors compared with nonmetastatic tumors. Higher expression levels in the metastatic compared with the nonmetastatic group were confirmed in 3 (HOTAIR, NEAT-1, UCA) of the 6 transcripts analyzed by qPCR (Fig. 2A and B) and the expression of MEG-3 was found to be decreased in the nonmetastatic tissues, which was consistent with data from primary cancer compared with nonmalignant tissue, suggesting that the expression levels of these lncRNAs increased following tumor development and progression. These analyses demonstrated the reliability and the validity of the procedure, since most of these transcripts have been described as cancer- or metastasis-associated genes. For instance, increased expression of lncRNA HOTAIR has previously been shown to be associated with metastasis in breast cancer patients, having a unique association with patient prognosis (8). Subsequently, HOTAIR expression level has been found to correlate with metastasis in colorectal carcinoma (9). Consistent with previous studies, MEG-3 expression was also downregulated in the metastatic compared with the nonmetastatic group (Fig. 2B).
Correlation of lncRNA expression with gender and age
We compared the expression of lncRNA in different gender and age groups. No statistically significant difference in gene expression was found between males and females. Similarly, no statistically significant difference was found between the younger and elder patients, after the classification of patients in different age groups based on the median age (50 years; data not shown).
Detection of lncRNA in salivary samples of OSCC patients
It has been previously shown that a subset of the salivary mRNAs can be used as biomarkers for oral cancer detection (31,32). To discover whether lncRNAs are present in salivary fluid, whole saliva was used for lncRNA identification.
In a pilot experiment, 1 ml salivary samples obtained from patients with OSCC (n=9), in which there were 4 patients with lymph node metastasis, were examined using qPCR for the presence of lncRNAs. Samples with a triplicate lncRNA qPCR result of Ct<40 were considered to be positive. Of the lncRNAs investigated, MALAT-1 was present in all the participants, while the mRNA expression was not significantly different between metastatic and nonmetastatic samples. However, HOTAIR was detected in 5/9 patients, 3 were positive of the 4 patients with lymph node metastasis, which is higher compared with those without metastasis. There was no detectable expression for the other lncRNAs investigated (Table III). Taken together, these data indicate that whole saliva contains detectable amounts of certain lncRNAs which may be potential markers for OSCC diagnosis.
Discussion
lncRNAs are a novel class of RNA molecules, >200-nucleotide long, with no protein-coding capacity (33). It has been found that several lncRNAs are able to control transcriptional alteration, implying that the difference of lncRNA profiling between normal and cancer cells is not merely the secondary effect of cancer transformation and lncRNAs are strongly associated with cancer progression (19). Thus, differential expression of lncRNAs may be profiled to aid cancer diagnosis, prognosis and select potential therapeutics.
In this study, we firstly confirmed that a subsets of lncRNAs were differentially overexpressed or underexpressed between tumor and matched adjacent non-tumor tissues. Specifically, HOTAIR, NEAT-1 and UCA1 were expressed at higher levels in tumors that subsequently metastasized than in others while expression of MEG-3 was downregulated. It is notable that we found no association between the expression of these lncRNAs with patients' gender and age, suggesting that lncRNA is a potential independent risk factor and diagnostic biomarker of OSCC.
Previous studies showed that salivary mRNAs can be used as biomarkers for oral cancer and the combined measurement of seven different mRNAs showed a specificity and sensitivity for oral cancer discrimination (18,31).
However, in the present study, only HOTAIR was detected in saliva with a statistically significant difference compared with the other lncRNAs investigated, particularly in samples from patients with lymph node metastasis. Metastasis is known to be the main cause of mortality from OSCC (34). Therefore, HOTAIR may potentially be used to predict patient survival. Furthermore, a large amount of samples need to be examined to determine the diagnostic value of HOTAIR in OSCC. In this study, despite the fact that MALAT-1 was detected in saliva, no significant difference between metastatic and nonmetastatic tissues was observed. This is in contrast to the well-documented lncRNA MALAT-1 association with metastasis. A possible reason for this is that the samples examined in this study were from different ethnic origin compared with the samples from other studies.
Comparable to, although on average less than, that of mRNAs, most lncRNAs have an extremely high stability (half-life >16 h), especially intergenic and cis-antisense RNAs, which are more stable compared with those derived from introns (35). Similar to intergenic and antisense RNA, HOTAIR may be a appropriate marker for OSCC. Gibb et al(36) reported that the most abundantly expressed lncRNA in normal oral mucosa was NEAT-1. However, in our experiments there was no detectable amount of NEAT-1 in saliva. It has been noted that NEAT-1 is one of the least stable lncRNAs (35). This may explain why there is no detectable expression in saliva despite the abundant amount of NEAT-1 in metastatic tissues.
In conclusion, we have shown that certain lncRNAs are present in whole saliva. One of these lncRNAs, HOTAIR, is differentially expressed in the saliva of the OSCC metastatic patients compared with that of primary cancer controls. These findings suggest that the detection of lncRNAs in saliva may be used as a noninvasive and rapid diagnostic tool for the diagnosis of oral cancer. Gibb et al(36) recently reported the first lncRNA expression map for the human oral mucosa. Furthermore, screening a large cohort of saliva from OSCC patients would determine the usefulness of differentially expressed lncRNAs as potential oral cancer biomarkers.
Acknowledgements
The study was supported by the Research Grants of Shenzhen Science and Technology Project (ZYA201106080030A), Fundamental Research Funds for the Central Universities of China (no. 21610404), the Scientific Research Foundation of Jinan University (no. 50624023), the Medical Science and Technology Foundation of Guangdong, the National Natural Science Foundation of China (no. 81102510) and the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry. The authors would like to thank Shenzhen Biomedical Research Support Platform and Shenzhen Public Service Platform for Molecular Diagnosis of Dermatology for their technical assistance.
Abbreviations:
|
OSCC |
oral squamous cell carcinoma |
|
ncRNAs |
non-coding RNAs |
|
lncRNAs |
long non-coding RNAs |
|
H&E |
hematoxylin and eosin |
References
|
de Camargo Cancela M, Voti L, Guerra-Yi M, Chapuis F, Mazuir M and Curado MP: Oral cavity cancer in developed and in developing countries: population-based incidence. Head Neck. 32:357–367. 2010.PubMed/NCBI | |
|
Ribeiro KC, Kowalski LP and Latorre MR: Impact of comorbidity, symptoms, and patients' characteristics on the prognosis of oral carcinomas. Arch Otolaryngol Head Neck Surg. 126:1079–1085. 2000. View Article : Google Scholar | |
|
Brosnan CA and Voinnet O: The long and the short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Mattick JS: Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Schöler N, Langer C, Döhner H, Buske C and Kuchenbauer F: Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp Hematol. 38:1126–1130. 2010.PubMed/NCBI | |
|
Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA and Jiang F: Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 67:170–176. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Xing L, Todd NW, Yu L, Fang H and Jiang F: Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 23:1157–1164. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S and Chang HY: Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S and Mori M: Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T, Tsunoda T, Daigo Y, Matsuda K, Kamatani N, Nakamura Y and Kubo M: Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 102:245–252. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 18:1243–1250. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 29:1810–1816. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M and Croce CM: Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP and Wei WI: Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E and Wong DT: Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 15:5473–5477. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW and Lin SC: Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 16:360–364. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gorenchtein M, Poh CF, Saini R and Garnis C: MicroRNAs in an oral cancer context - from basic biology to clinical utility. J Dent Res. 91:440–446. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Huarte M and Rinn JL: Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gibb EA, Brown CJ and Lam WL: The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI | |
|
Prensner JR and Chinnaiyan AM: The emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Tinzl M, Marberger M, Horvath S and Chypre C: DD3PCA3 RNA analysis in urine - a new perspective for detecting prostate cancer. Eur Urol. 46:182–186. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, Yamamoto D, Alvarenga R, da Silva CC, Brito GC, Simon SD, Moreira-Filho CA, Leite KR, Camara-Lopes LH, Campos FS, Gimba E, Vignal GM, El-Dorry H, Sogayar MC, Barcinski MA, da Silva AM and Verjovski-Almeida S: Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 23:6684–6692. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H and Muller-Tidow C: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Kim YS, Hwan JD, Bae S, Bae DH and Shick WA: Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 10:5762010. View Article : Google Scholar : PubMed/NCBI | |
|
Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H and Akkawi M: Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 21:688–692. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, Zhang Y, Xin DQ, Na YQ and Chen WF: Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T and Noguchi M: Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 97:106–112. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Lin R, Maeda S, Liu C, Karin M and Edgington TS: A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH and Wong DT: Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 10:8442–8450. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Park NJ, Li Y, Yu T, Brinkman BM and Wong DT: Characterization of RNA in saliva. Clin Chem. 52:988–994. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Lipovich L, Johnson R and Lin CY: MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Scapoli L, Palmieri A, Lo ML, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and Carinci F: MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 23:1229–1234. 2010.PubMed/NCBI | |
|
Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME and Mattick JS: Genome-wide analysis of long noncoding RNA stability. Genome Res. 22:885–898. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Gibb EA, Enfield KS, Stewart GL, Lonergan KM, Chari R, Ng RT, Zhang L, MacAulay CE, Rosin MP and Lam WL: Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral Oncol. 47:1055–1061. 2011. View Article : Google Scholar : PubMed/NCBI |