Whole exome sequencing identified a pathogenic nonsense mutation in LMNA in a family with a progressive cardiac conduction defect: A case report
- Authors:
- Published online on: April 1, 2020 https://doi.org/10.3892/mmr.2020.11048
- Pages: 2459-2465
-
Copyright: © Fan et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Progressive cardiac conduction defect (PCCD), also known as Lenegre-Lev disease (Online Mendelian Inheritance in Man entry no. 113900), is a rare bradycardic disorder (1,2). PCCD is characterized by electrical deterioration of the conduction system in the atrium and ventricle, which presents as temporal prolongation of the PR interval, bradycardia, bundle branch block and advanced atrioventricular block (AVB) (3). A genetic epidemiological study reported that the PCCD frequency ranged from 0.21–2.28% in >6,600 patients implanted with a pacemaker in western France (4). In the past several decades, the rapid development of molecular genetics has provided important data on the genetic basis of PCCD (5).
Clinical and molecular studies have led to the characterization of PCCD as an inherited disease (6). Several genes have been associated with PCCD, including sodium voltage-gated channel α5, protein kinase AMP-activated non-catalytic subunit γ2, NK2 homeobox 5 and lamin A/C (LMNA) (5,7). LMNA is located on chromosome 1q21.2–21.3 and generates both lamin A and C by alternative splicing of a single transcript (8). Lamin A and C are nuclear intermediate filament proteins that form one of the major structural components of the lamina network, which underlies and mechanically supports the nuclear envelope (9). LMNA mutations cause a spectrum of multisystem diseases, including PCCD, dilated cardiomyopathy (DCM), muscular dystrophy and lipodystrophy (6,9,10). Although pleiotropic phenotypes are expressed in LMNA-associated diseases, PCCD is one of the most malignant phenotypes, with an untreated sudden cardiac death rate of ≤46% (11). Mortality occurred in 8% of LMNA genotype-positive subjects at a mean age of 66±8 years, over a mean follow-up period of 7.8±6.3 years (12). These findings illustrate the importance of genetic screening and genetic counseling in patients with PCCD and families with genetic mutations.
In the present study, whole-exome sequencing of three patients with inherited PCCD and one healthy control subject was performed, and a pathogenic mutation was identified. The PCCD-associated mutation was identified as a nonsense LMNA mutation, which truncates the LMNA protein. Family screening identified LMNA genotype-positive carriers, potentially allowing for improved management of mutation carriers in the future.
Case report
Patients and DNA samples
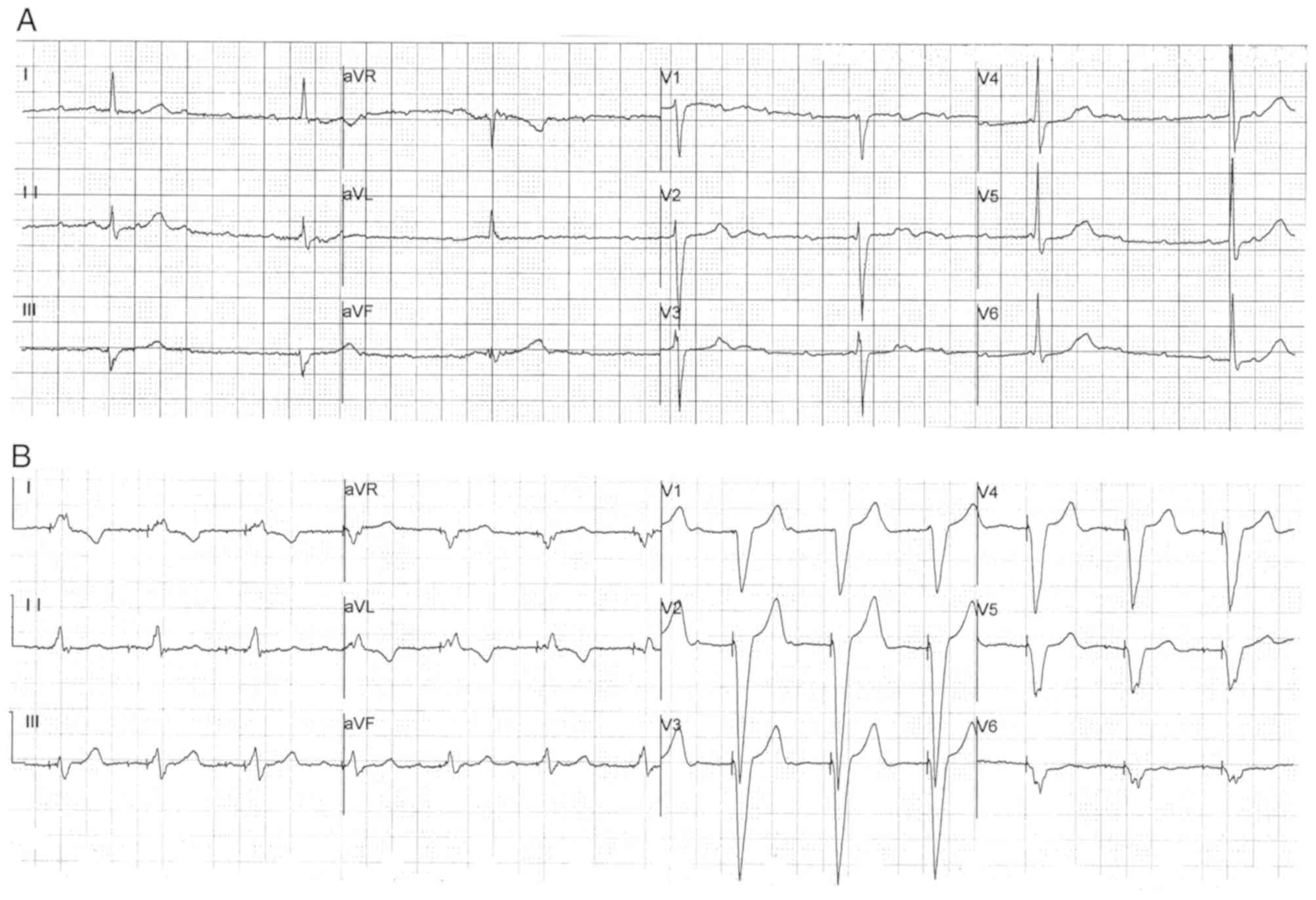
The proband (III-4), a 57 year-old Chinese woman, initially presented with chest discomfort, repeated attacks of dizziness and amaurosis. The patient presented with the aforementioned symptoms 12 years ago (in 2000) and received no treatment prior to their first hospitalization in 2012. Due to persistent symptoms, the patient was admitted to Department of Cardiology in Fuwai Hospital (July 2012) for comprehensive diagnosis and treatment. The 24 h ambulatory electrocardiogram (ECG) results identified sinus bradycardia [average heart rate, 48 beats per minute (bpm); minimum heart rate, 23 bpm] and advanced AVB in the patient (Fig. 1A). An echocardiogram identified an enlargement of the left atrium (left atrial diameter, 43 mm). Therefore, the patient received a permanent pacemaker with a normal pacing ECG (Fig. 1B), which alleviated the aforementioned symptoms. By investigation of the familial cardiac history of the patient, a further ten relatives also experiencing dizziness, syncope and bradycardia were identified. Among these symptomatic family members, six participants had an implanted pacemaker (II-10 also had an implantable cardioverter defibrillator), and four participants experienced sudden death. In addition, other common cardiovascular diseases were excluded in the family, including coronary heart disease, DCM and myocarditis. The direct family history, including cases of bradycardia, pacemaker implantation and sudden death, raised suspicion of an inherited arrhythmia disease.
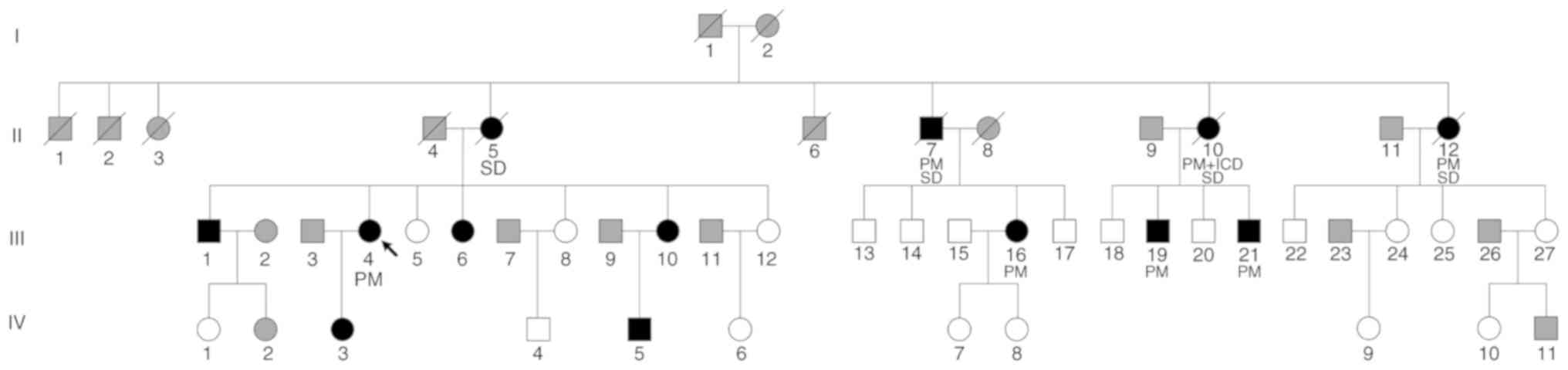
A total of 39 participants (male patients, 10; female patients, 29; age range, 13–68 years; mean age, 42.59 years) from a Chinese family were included in Department of Cardiology in Fuwai Hospital (September 2017). The pedigree chart is presented in Fig. 2. Each participant provided peripheral venous blood for biochemical and genetic analyses and was examined by an ECG and echocardiography. A clinical survey of the cardiac disease history of each participant was also performed. Then, 2 ml anticoagulated venous blood samples from each participant were contained in EDTA vacuum tubes. Genomic DNA was extracted from each blood sample using the QIAamp DNA Blood Mini kit (Qiagen GmbH), according to the manufacturer's instructions.
The present study was approved by the Ethics Committee of Fuwai Hospital. Written informed consent was obtained from each participant.
Whole-exome sequencing (WES)
WES was performed on blood samples from three participants with PCCD (III-4, III-16 and III-21) and a healthy family member (III-5) from the same generation of the family using an Illumina HiSeq 2000 sequencing system (Illumina, Inc.). The average coverage depth of the WES was >200-fold. The SureSelect Human All Exon V5 (Agilent Technologies, Inc.) was used for exome capture and Burrows-Wheeler Aligner (version 0.7.12) (13) was used to align the reads to the National Center for Biotechnology Information human reference genome (GRCh37/hg19) (https://www.ncbi.nlm.nih.gov/projects/genome/guide/human/index.shtml), after removing sequence adaptors and low-quality reads (the proportion of reads with undetermined base information in single-end sequencing >10%; single read >50% low quality (<5) nucleotides). Picard (version 1.119; sourceforge.net/projects/picard) and Genome Analysis Toolkit (version 3.7) (14) tools were used for duplicate removal, local realignment and base quality recalibration. Single nucleotide polymorphism (SNP) and Indel calling were performed using Sequence Alignment Map tools (version 1.10; samtools.sourceforge.net/) and were annotated using Annotate Variation (version 20170825) (15), according to the SNP database (www.ncbi.nlm.nih.gov/SNP) and the 1000 Genomes Project database (www.1000genomes.org/index.html).
Genotype identification
Firstly, all potentially significant variants were identified by in silico analysis, using the Protein Variation Effect Analyzer (version 1.1.5; provean.jcvi.org/index.php), Polymorphism Phenotyping (version 2.0; genetics.bwh.harvard.edu/pph2), Sorting Intolerant From Tolerant (version 4.0.3; sift.jcvi.org) and Mutation Taster (version 2.0; www.mutationtaster.org) bioinformatics tools. The conservation status of the candidate variants was estimated by the Genome Evolutionary Rate Processing (version 2.0; mendel.stanford.edu/sidowlab/downloads/gerp/index.html), Phylogenetic Analysis with Space/Time Models (PHAST) Cons (version 1.3; compgen.bscb.cornell.edu/phast) and PhyloP (version 1.3; compgen.bscb.cornell.edu/phast/help-pages/phyloP.txt) software.
In addition, Sanger sequencing was used to identify the validity of the variants. PCR of every potential variant was amplified DNA from the proband. A primer pair was used to amplify the LMNA variant c.1443C>A (forward, 5′-AGATGGAAGGAGAGGCCTCA-3′ and reverse, 5′-AGAAAGGGGCCCTGAATTGG-3′). PCR was performed with a TaKaRa LA Taq kit with GC buffer (cat. no. RR02AG; Takara Biomedical Technology Co., Ltd.) according to manufacturer's protocol. The amplification process was: Initial denaturation for 1 min at 94°C, followed by 30 cycles of 30 sec at 94°C, 30 sec at 60°C, 2 min at 72°C; and a final extension step at 72°C for 5 min. Subsequently, the candidate LMNA variants were sequenced in other family members and 100 healthy controls of matched ancestry (without any cardiovascular disease). The control group (male patients, 53; female patients, 47; age range, 18–60 years; mean age, 40.35 years) was recruited from September 2016 to August 2017. The PCR products were sequenced using an ABI Prism® 377 DNA sequencer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The identification of the variants was performed using Chromas software (version 2.22; Technelysium Pty, Ltd.). The co-segregation of genotype and phenotype was analyzed.
Results
Clinical examinations
The ECG examinations identified similar symptoms among the participants in terms of bradycardia, extended PR interval and advanced AVB (Table I). The most common characteristic was bradycardia, which was observed in 12 patients from the family. The minimum heart rate recorded for each participant was ≤50 bpm. An extended PR interval >200 msec was recorded in seven participants (III-1, III-4, III-6, III-10, III-16, III-19 and III-21). A cardiac conduction block manifested in different forms, including a right bundle branch block, an intraventricular block and various degrees of AVB. Advanced AVB was identified in nine patients, of which eight (except III-1) required the implantation of a pacemaker. Moreover, II-10 had also received an implantable cardioverter defibrillator to prevent sudden death. However, one identified mutation carrier (IV-3) displayed no abnormal ECG findings, and although individual IV-5 presented with bradycardia, the PR interval in the ECG was within the normal range.
None of the participants displayed hypertension or muscle weakness. Investigation of serum creatinine phosphokinase displayed no abnormalities in each participant. There was no evidence supporting the diagnosis of DCM, although several patients displayed abnormalities in the echocardiograph. Participants III-1 and III-19 had enlarged hearts, mitral and tricuspid insufficiency and a normal ejection fraction. Participant III-6 had an atrial septal defect and right atrial and ventricular enlargement (right ventricular anteroposterior diameter, 39 mm). After repair of the atrial septal defect, the right ventricular anteroposterior diameter decreased to 21 mm. Participant III-16 had a mild enlargement of the left heart due to mitral and tricuspid insufficiency. Participant III-21 suffered from patent foramen ovale. Other mutation carriers (III-10, IV-3 and IV-5) displayed a normal echocardiograph.
Identification of pathogenic mutations
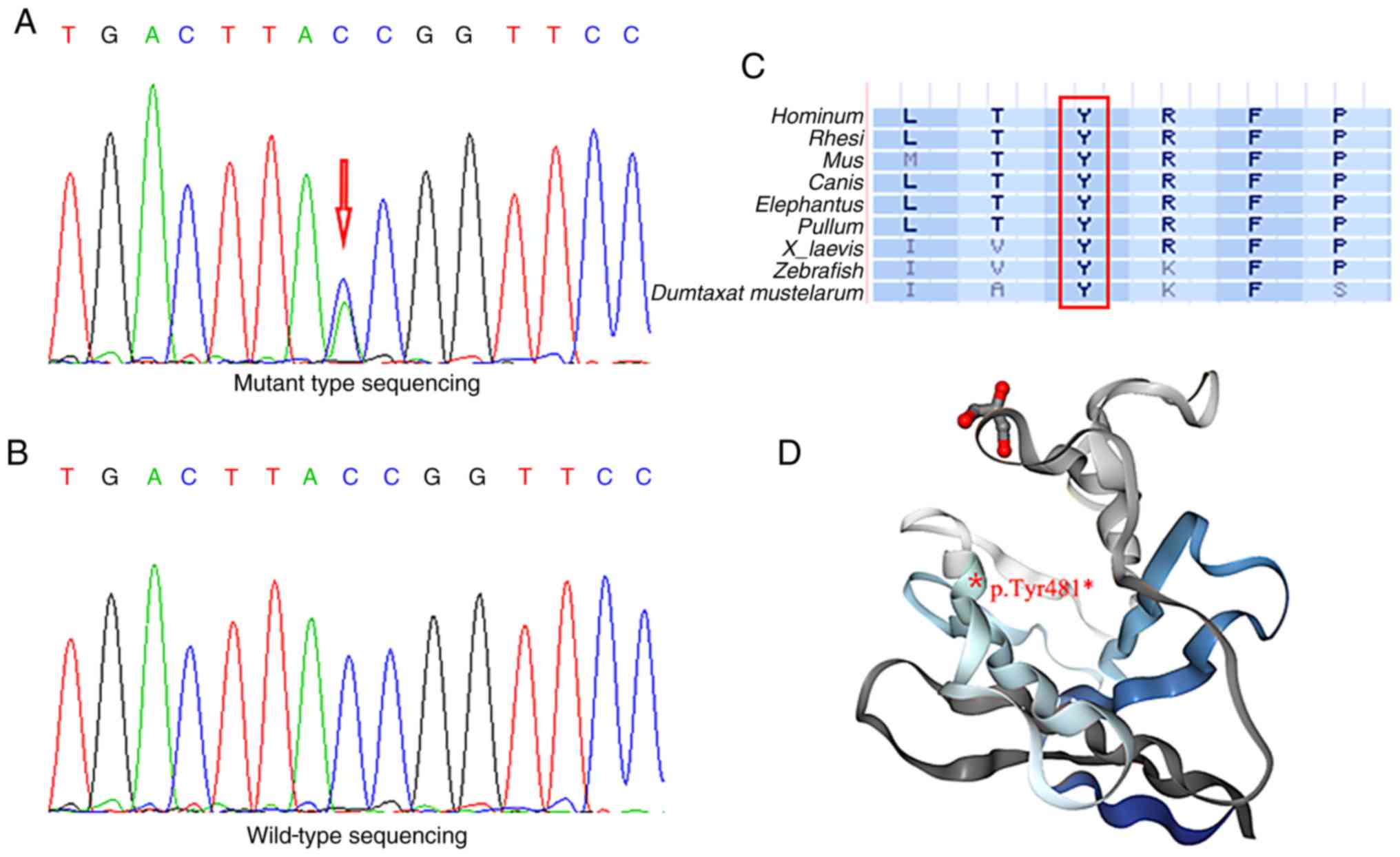
Whole-exome sequencing was performed in three participants (III-4, III-16 and III-21) and a healthy family member (III-5). A total of 5,401 non-synonymous variants were detected in the exonic and untranslated regions after filtering by frequency <1% from the 1000 Genomes Project database. Co-association analysis identified six candidate genes (LMNA, ARFGEF3, FNDC1, MAN1A2, ODF2L and YY1AP1) associated with cardiac disease. Furthermore, Sanger sequencing identified all possible pathogenic variants among all participants. A nonsense LMNA mutation p.Tyr481* (Fig. 3A) was identified as a pathogenic mutation in the patients with PCCD. The genotype and phenotype were only completely co-associated for this variant in the family included in the present study. In 100 healthy controls, the same mutation was not detected (Fig. 3B). To investigate whether the LMNA mutation caused significant alterations to gene and protein expression, several bioinformatics tools were employed (Table II). The nucleotide at position 1,443 and the amino acid at position 481 were fully conserved in nine vertebrates (Fig. 3C). To predict the functional effects of the LMNA mutation, in silico analyses were conducted and suggested that the nonsense mutation was a risk factor for PCCD. It was further suggested that the nonsense LMNA mutation generated a truncated LMNA protein, lacking the final 184 amino acids (Fig. 3D).
Discussion
In the present study, a large Chinese family with inherited PCCD were investigated and an LMNA mutation, p.Tyr481*, was identified by whole-exome sequencing. The present study suggested that the LMNA mutation played a crucial role in the genetic pathogenesis of PCCD in the aforementioned family. Genetic testing of LMNA offers the possibility for hereditary diagnosis of patients with PCCD and the opportunity to take precautionary actions to prevent sudden death as early as possible (16).
As a hereditary syndrome, PCCD is related to genetic variants in multiple genes, including LMNA (5,17). At present, a large number of disease-causing LMNA mutations have been described (16,18). The LMNA mutations are predominantly missense mutations, but nonsense, frameshift and intragenic deletions and duplications have also been reported (16). Non-missense mutations in LMNA have an influence on the pathogenesis of PCCD (19). Nishiuchi et al (20) reported that truncation mutation carriers presented a worse prognosis than those with a missense mutation, based on gene risk stratification of cardiac disorders in 77 LMNA mutation carriers from 45 Japanese families. van Rijsingen et al (21) evaluated risk factors for malignant ventricular arrhythmias in a multicenter cohort of 269 LMNA mutation carriers, and reported that non-missense mutations were an independent risk factor for malignant ventricular arrhythmias. In the present study, the participants had a high risk of advanced AVB and six subjects (II-10, II-12, III-4, III-16, III-19 and III-21) were implanted with a pacemaker. Therefore, regarding the family investigated in the present study, it could be suggested that other mutation carriers should be kept under careful observation.
LMNA mutations are associated with several cardiac phenotypes, including cardiac conduction disorders, DCM, malignant ventricular arrhythmia and left ventricular dysfunction (17,22). The most prominent cardiac feature of LMNA mutations is PCCD (23). The majority of the participants in the present study presented with dizziness and chest discomfort and suffered from bradycardia on initial ECG examination. As the disease progressed with age, certain participants developed advanced AVB and required the implantation of a pacemaker. Nevertheless, clinical variability has been observed among mutation carriers with the same LMNA mutation in present study. The age of onset for the majority of the participants in the present study was ~40 years. Participants IV-3 (age, 13 years) and IV-5 (age, 18 years) did not present with typical symptoms, potentially due to their young age. Therefore, it may be important for these individuals to attend future follow-up appointments.
Variable phenotypes in LMNA mutations may be caused by factors, including gender, onset age and mutation location (16). A number of studies have reported significant gender differences in cardiac phenotypes (17,21,24), and it has been further reported that estrogen may play a role in protecting against cardiac dysfunction (25,26). Ollila et al (22) reported that male LMNA mutation carriers have an earlier onset of disease than females. However, no gender differences in the cardiac phenotypes were identified in the family included in the present study. In addition, patients with LMNA mutations display age-dependent differences in the manifestation of cardiac phenotypes and prognosis (27). Consistent with the literature, in the present study, the greater the age of the patient with PCCD, the more severe the cardiac phenotype and prognosis (16,27). Furthermore, the LMNA mutation location can predict the severity of the cardiac phenotype (16). LMNA mutations upstream of the nuclear localization signal or the C-terminal tail domain are associated with a more severe cardiac phenotype overall than those occurring downstream (16). The mutation identified in the present study, p.Tyr481*, was located in the C-terminal tail domain of LMNA, which is consistent with other severe cardiac phenotypes (16,17).
In the same codon at position 481, other nucleotide mutant forms and phenotypes have been reported (24,28–32). A missense mutation (c.1442A>G, p.Tyr481Cys) was detected in patients with DCM by clinical DNA sequencing (28–30). Kitaguchi et al (31) first reported a case of limb-girdle muscular dystrophy with atrioventricular conduction block resulting from an LMNA missense mutation (c.1441T>C, p.Tyr481His). In terms of nonsense mutations, Sylvius et al (32) identified a proband with DCM carrying a heterozygous mutation (c.1443C>G, p.Tyr481*). Furthermore, in a multicenter cohort study of 269 LMNA mutation carriers, the same mutation (c.1443C>A, p.Tyr481*) was identified without detailed data of phenotypes or family history (24). The present study, which provided clinical and genetic data for a large family with PCCD, has enriched the association of genotype and phenotype for LMNA mutations.
Early-onset PCCD in a structurally normal heart should prompt consideration of PCCD genetic testing, particularly if there is a family history of PCCD, pacemaker implantation or sudden death (33). Once a proband with a known pathogenic LMNA mutation has been screened, it is necessary to perform a family screening to identify other carriers of the mutation (34). For relatives of a sudden death victim with an unknown genotype, family screening for an LMNA mutation is a class IIa level of recommendation, based on the 2015 European Society of Cardiology Guidelines for the Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death (34). The number of asymptomatic relatives identified with LMNA mutations and subclinical phenotypes is increasing (35). Family screening distinguished genotype positives and negatives in the family included in the present study. Individuals IV-3 and IV-5 were genotype positive but did not present with typical symptoms or advanced AVB, therefore it could be suggested that these individuals should be offered life-long follow-up care (36).
In conclusion, the present study suggested that the c.1443C>A, p.Tyr481* mutation in LMNA may cause PCCD. However, further investigation is required to fully understand the functional changes of this mutation. Family genetic counseling and sequencing are vital to enable early identification of LMNA genotype-positive family members, risk stratification and determination of therapeutic timing. Further functional identification would be beneficial for improving the existing knowledge of the pathogenesis of PCCD.
Acknowledgements
Not applicable.
Funding
The present study was supported by the CAMS Innovation Fund for Medical Sciences (grant no. 2016-I2M-1-002), the National Key Research and Development Program of China (grant no. 2016YFC1300100), the National Natural Science Foundation of China (grant no. 81600305), the Beijing Municipal Science and Technology Commission (grant no. Z151100003915078), the PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (grant nos. 3332015108, 3332015205, 3332018058 and 3332019134), the PUMC Graduate Innovation Fund (grant no. 2018-1002-01-14).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LPW and XLZ designed and supervised the research. PF, DZ and KQY collected the samples and the clinical information. PF, DZ and TT performed the experiments. PF, KQY, FL and YXL performed the data analysis. PF wrote the manuscript. YXL, LPW and XLZ edited the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Fuwai Hospital. Each participant provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Lenegre J and Moreau P: Chronic auriculo-ventricular block. Anatomical, clinical and histological study. Arch Mal Coeur Vaiss. 56:867–888. 1963.(In French). PubMed/NCBI | |
|
Lev M, Kinare SG and Pick A: The pathogenesis of atrioventricular block in coronary disease. Circulation. 42:409–425. 1970. View Article : Google Scholar : PubMed/NCBI | |
|
Kawaguchi T, Hayashi H, Miyamoto A, Yoshino T, Taniguchi A, Naiki N, Sugimoto Y, Ito M, Xue Q, Murakami Y and Horie M: Prognostic implications of progressive cardiac conduction disease. Circ J. 77:60–67. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Gourraud JB, Kyndt F, Fouchard S, Rendu E, Jaafar P, Gully C, Gacem K, Dupuis JM, Longueville A, Baron E, et al: Identification of a strong genetic background for progressive cardiac conduction defect by epidemiological approach. Heart. 98:1305–1310. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Baruteau AE, Probst V and Abriel H: Inherited progressive cardiac conduction disorders. Curr Opin Cardiol. 30:33–39. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Rankin JS and Ellard S: The laminopathies: A clinical review. Clin Genet. 70:261–274. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Rogozhina Y, Mironovich S, Shestak A, Adyan T, Polyakov A, Podolyak D, Bakulina A, Dzemeshkevich S and Zaklyazminskaya E: New intronic splicing mutation in the LMNA gene causing progressive cardiac conduction defects and variable myopathy. Gene. 595:202–206. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Lin F and Worman HJ: Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 268:16321–16326. 1993.PubMed/NCBI | |
|
Mattout A, Dechat T, Adam SA, Goldman RD and Gruenbaum Y: Nuclear lamins, diseases and aging. Curr Opin Cell Biol. 18:335–341. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Mestroni L and Taylor MR: Lamin A/C gene and the heart: How genetics may impact clinical care. J Am Coll Cardiol. 52:1261–1262. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
van Berlo JH, de Voogt WG, van der Kooi AJ, van Tintelen JP, Bonne G, Yaou RB, Duboc D, Rossenbacker T, Heidbuchel H, de Visser M, et al: Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: Do lamin A/C mutations portend a high risk of sudden death? J Mol Med (Berl). 83:79–83. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Hasselberg NE, Haland TF, Saberniak J, Brekke PH, Berge KE, Leren TP, Edvardsen T and Haugaa KH: Lamin A/C cardiomyopathy: Young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 39:853–860. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Li H and Durbin R: Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wang K, Li M and Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI | |
|
Captur G, Arbustini E, Syrris P, Radenkovic D, O'Brien B, McKenna WJ and Moon JC: Lamin mutation location predicts cardiac phenotype severity: Combined analysis of the published literature. Open Heart. 5:e0009152018. View Article : Google Scholar : PubMed/NCBI | |
|
Kawakami H, Ogimoto A, Tokunaga N, Nishimura K, Kawakami H, Higashi H, Iio C, Kono T, Aono J, Uetani T, et al: A novel truncating LMNA mutation in patients with cardiac conduction disorders and dilated cardiomyopathy. Int Heart J. 59:531–541. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Captur G, Arbustini E, Bonne G, Syrris P, Mills K, Wahbi K, Mohiddin SA, McKenna WJ, Pettit S, Ho CY, et al: Lamin and the heart. Heart. 104:468–479. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Priori SG, Blomstrom-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J, Elliott PM, Fitzsimons D, Hatala R, Hindricks G, et al: 2015 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The task force for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death of the european society of cardiology (ESC). Endorsed by: Association for European paediatric and congenital cardiology (AEPC). Eur Heart J. 36:2793–2867. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Nishiuchi S, Makiyama T, Aiba T, Nakajima K, Hirose S, Kohjitani H, Yamamoto Y, Harita T, Hayano M, Wuriyanghai Y, et al: Gene-based risk stratification for cardiac disorders in LMNA mutation carriers. Circ Cardiovasc Genet. 10:e0016032017. View Article : Google Scholar : PubMed/NCBI | |
|
van Rijsingen IA, Arbustini E, Elliott PM, Mogensen J, Hermans-van Ast JF, van der Kooi AJ, van Tintelen JP, van den Berg WP, Pilotto A, Pasotti M, et al: Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 59:493–500. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Ollila L, Nikus K, Holmstrom M, Jalanko M, Jurkko R, Kaartinen M, Koskenvuo J, Kuusisto J, Karkkainen S, Palojoki E, et al: Clinical disease presentation and ECG characteristics of LMNA mutation carriers. Open Heart. 4:e0004742017. View Article : Google Scholar : PubMed/NCBI | |
|
Brodt C, Siegfried JD, Hofmeyer M, Martel J, Rampersaud E, Li D, Morales A and Hershberger RE: Temporal relationship of conduction system disease and ventricular dysfunction in LMNA cardiomyopathy. J Card Fail. 19:233–239. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
van Rijsingen IA, Nannenberg EA, Arbustini E, Elliott PM, Mogensen J, Hermans-van Ast AJ, van der Kooi AJ, van Tintelen JP, van den Berg MP, Grasso M, et al: Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur J Heart Fail. 15:376–384. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Du XJ: Gender modulates cardiac phenotype development in genetically modified mice. Cardiovasc Res. 63:510–519. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Babiker FA, De Windt LJ, van Eickel M, Grohe C, Meyer R and Doevendans PA: Estrogenic hormone action in the heart: Regulatory network and function. Cardiovasc Res. 53:709–719. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Nakajima K, Aiba T, Makiyama T, Nishiuchi S, Ohno S, Kato K, Yamamoto Y, Doi T, Shizuta S, Onoue K, et al: Clinical manifestations and long-term mortality in lamin A/C mutation carriers from a Japanese multicenter registry. Circ J. 82:2707–2714. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, et al: Reassessment of mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 19:192–203. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ito K, Patel PN, Gorham JM, McDonough B, De Palma SR, Adler EE, Lam L, MacRae CA, Mohiuddin SM, Fatkin D, et al: Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc Natl Acad Sci U S A. 114:7689–7694. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, et al: The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 16:601–608. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Kitaguchi T, Matsubara S, Sato M, Miyamoto K, Hirai S, Schwartz K and Bonne G: A missense mutation in the exon 8 of lamin A/C gene in a Japanese case of autosomal dominant limb-girdle muscular dystrophy and cardiac conduction block. Neuromuscul Disord. 11:542–546. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Sylvius N, Bilinska ZT, Veinot JP, Fidzianska A, Bolongo PM, Poon S, McKeown P, Davies RA, Chan KL, Tang AS, et al: In vivo and in vitro examination of the functional significances of novel lamin gene mutations in heart failure patients. J Med Genet. 42:639–647. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, et al: HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: This document was developed as a partnership between the heart rhythm society (HRS) and the European heart rhythm association (EHRA). Heart Rhythm. 8:1308–1339. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Priori SG, Blomstrom-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J, Elliott PM, Fitzsimons D, Hatala R, Hindricks G, et al: 2015 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Rev Esp Cardiol (Engl Ed). 69:1762016.PubMed/NCBI | |
|
Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Bohm M, Duboc D, Gimeno J, de Groote P, Imazio M, et al: Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: A position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 37:1850–1858. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Mogensen J, van Tintelen JP, Fokstuen S, Elliott P, van Langen IM, Meder B, Richard P, Syrris P, Caforio AL, Adler Y, et al: The current role of next-generation DNA sequencing in routine care of patients with hereditary cardiovascular conditions: A viewpoint paper of the European society of cardiology working group on myocardial and pericardial diseases and members of the European society of human genetics. Eur Heart J. 36:1367–1370. 2015. View Article : Google Scholar : PubMed/NCBI |