Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma
- Authors:
- Published online on: December 28, 2012 https://doi.org/10.3892/or.2012.2219
- Pages: 946-950
Abstract
Introduction
Recently, many studies have focused on investigating the role of non-coding RNAs (ncRNAs), RNA sequences that are not translated into protein. Not only are the functions of ncRNAs only partially understood, but the clinical or biological significance of most ncRNAs has not yet been determined. Among ncRNAs, long ncRNAs, consisting of over 200 bases have been reported to associate with DNA-binding proteins, such as chromatin modifying complexes, and certain ncRNAs can epigenetically regulate the expression of multiple genes through this mechanism (1–3).
Hox transcript antisense intergenic RNA (HOTAIR) is a long ncRNA that was identified from a custom tilling array of the HOXC locus (12q13.13) (4). HOTAIR forms a complex with the polycomb-repressive complex 2 (PRC2), composed of EZH2, SUZ12 and EED, to trimethylate histone H3 at lysine 27 (H3K27me3), thereby inhibiting HOXD gene expression. Thus, HOTAIR epigenetically regulates the expression of HOXD, a gene located on a separate chromosome. Recent studies have demonstrated the clinical significance of HOTAIR in surgical solid malignancies. Gupta et al found that patients with high HOTAIR expression in primary lesions had a poorer prognosis for both overall- and metastases-free survival than those with low HOTAIR expression in breast cancer (4). Moreover, we reported the clinical significance of HOTAIR in patients with advanced colorectal cancer (CRC), where HOTAIR was more highly expressed in cancerous tissues than in noncancerous tissues (5). High HOTAIR expression correlated well with the presence of liver metastases, and was significantly associated with poorer prognoses. In addition, we found that HOTAIR expression was associated with a genome-wide reprogramming of PRC2 (SUZ12, EZH2 and H3K27me3) by gene set enrichment analysis using cDNA array data from a pilot study (5). Since HOTAIR expression has clinical significance in both breast and colorectal cancers, we expect that this ncRNA may also play an important role in other intractable malignancies.
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world. In Japan, HCC is an intractable tumor, causing approximately 30,000 deaths per year and representing the third leading cause of death from malignant neoplasms in men, and the fifth leading cause of death from malignant neoplasms in women (6,7). The major causes of HCC are viral infections, alcohol and tobacco use. While efforts have been made to identify appropriate prognostic markers for HCC (8–12), including primary tumor size, elevated AFP levels, and gene expression markers in the primary tumor, these method have not proven adequate to predict the prognosis of all HCC patients. In addition, few studies describing the expression of ncRNAs and their clinical significance in HCC have been published. In the present study, we determined the clinicopathological significance of HOTAIR expression in HCC patients, and investigated whether HOTAIR is a useful prognostic indicator in HCC patients.
Materials and methods
Clinical tissue samples
A total of 64 patients with HCC who underwent surgery at Beppu Hospital were enrolled in this study. The resected tumor and paired non-tumor tissue specimens were immediately frozen in liquid nitrogen and kept at −80<C until analysis. Frozen tissue specimens were homogenized in guanidinium thiocyanate, and total RNAs were obtained by ultracentrifugation through a cesium chloride cushion. Written informed consent was obtained from all patients. All patients were closely followed after surgery at regular one-month intervals.
RNA preparation, reverse transcription and quantitative real-time PCR
Total RNA from frozen HCC samples (n=64) was extracted using Isogen (Nippon Gene Co., Ltd.) following the manufacturer’s protocol. As previously reported, cDNAs from all samples were synthesized from 8.0 μg of total RNA (13). HOTAIR levels were quantified using a LightCycler 480 Probes Master kit (Roche Applied Science) following the manufacturer’s protocol with the following specific HOTAIR primers: (forward, 5′-CAGTGGGGAACTCTGACTCG-3′ and reverse, 5′-GTGCCTGGTGCTCTCTTACC-3′). HOTAIR levels were normalized to GAPDH (forward, 5′-GTCAACGGATTTGG TCTGTATT-3′ and reverse, 5′-AGTCTTCTGGGTGGCAGT GAT-3′).
Cell lines
HepG2 cells, human liver cancer cells, were provided by the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University, Japan. All cell lines were maintained in Dulbecco’s modified Eagle’s media (DMEM) supplemented with 10% fetal calf serum and antibiotics. We cultured the cells at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
HOTAIR expression lentiviral vector
To generate a HOTAIR expression lentiviral vector, we amplified full-length human HOTAIR by PCR using MCF7 cDNA. Lentiviruses were produced by transient transfection of HEK293T cells with pCMV-VSV-G-RSV-Rev, pCAG-HIVgp, and either CSII-CMV-HOTAIR or CSII-CMV-MCS (empty) plasmid DNAs (5′-XhoI and 3′-NotI sites) using Lipofectamine 2000 (Invitrogen), following the manufacturer’s protocol. Forty-eight hours after cotransfection, the lentivirus-containing supernatant was collected and passed through a 0.45-μm filter. The titer of the lentivirus vector in filtered supernatants was estimated by measuring the concentration of HIV p24 gag antigen with an ELISA kit (Perkin-Elmer Life Science).
Cell proliferation assay
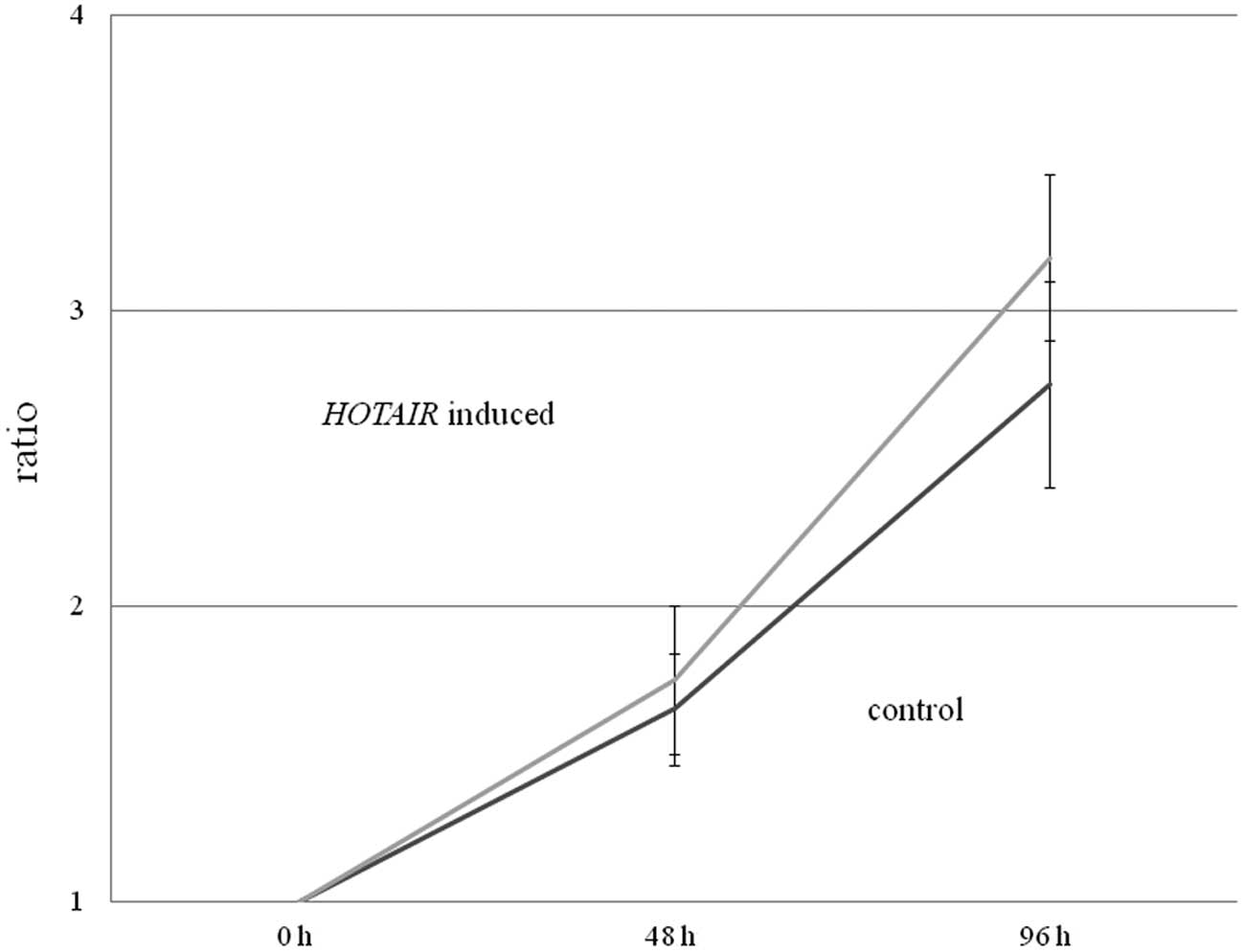
Cell proliferation was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. In brief, we plated HepG2 cells infected with either the human HOTAIR full-length lentiviral vector or the empty lentiviral vector, in 96-well tissue culture plates at a density of 5.0×103 cells per well. At different time points (24, 72, and 120 h after plating, representing the 0-, 48-, and 96-h time points respectively), 10 μl MTT (5 mg/ml in phosphate-buffered saline) was added to each well, and plates were incubated for an additional 4 h at 37°C. The colored formazan product was then dissolved in 100 μl DMSO. We then evaluated mitochondrial activity, reflecting cellular growth and viability, by measuring the optical density at a test wavelength of 570–650 nm using a microplate reader (Bio-Rad, Japan); results were expressed as optimal density per milligram protein (OD/mg protein).
Statistical analysis
The significance of differences between two groups was estimated using the Student’s t-test and the χ2 test. Overall survival curves were plotted according to the Kaplan-Meier method, with the log-rank test applied for comparison. Variables with a p-value of <0.05 by univariate analysis were used in subsequent multivariate analysis on the basis of the Cox proportional hazards model. All differences were considered statistically significant when p<0.05. Statistical analyses were conducted using JMP 5 software (SAS Institute).
Gene set enrichment analysis (GSEA) of HCC in the GEO database
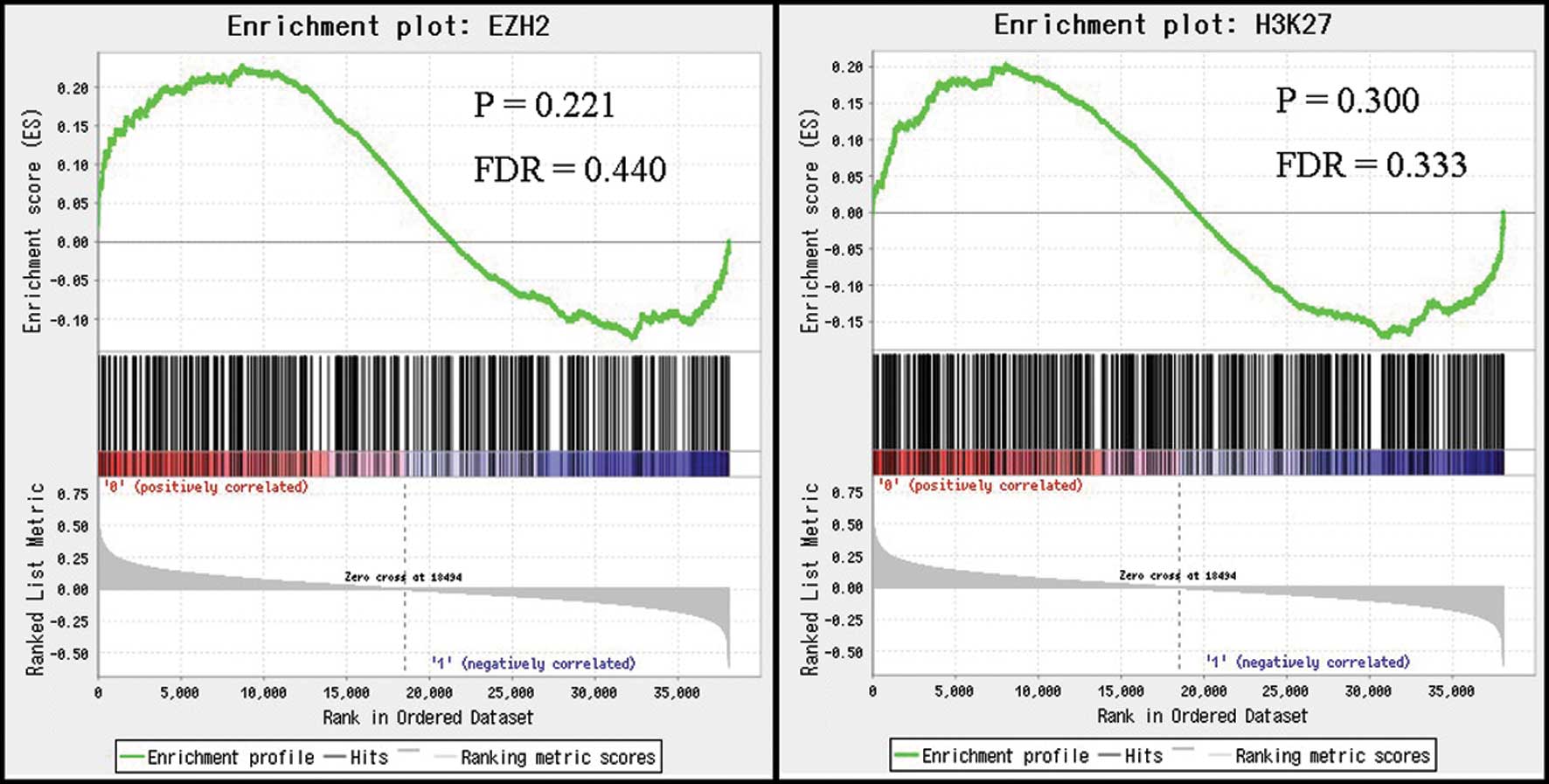
For GSEA (14), we used GSE27462 in the GEO database; this was a dataset for RNA expression profiles collected using a genome tiling array in 10 HCC samples (15). HOTAIR expression was treated as a binary variable divided into low or high expression according to medians. For functional gene sets for GSEA, we used gene sets arising from global occupancy of H3K27me3, and EZH2, a PRC2 subunit induced by HOTAIR overexpression in MDA-MB-231 breast cancer cells (4). As a metric for ranking genes in GSEA, the difference between the means of samples with low and high HOTAIR expression was used, and other parameters were set to default values.
Results
We first evaluated HOTAIR expression in primary tumors from HCC patients (n=64) by quantitative real-time PCR. From these data, we divided the 64 patients with HCC into a HOTAIR high expression group (n=13) and a low expression group (n=51) (Fig. 1A). In consideration of clinical applications, we set cut-off values as the upper limit of normal samples, and we found a HOTAIR/GAPDH ratio of 0.027 in HCC samples. Patients with high HOTAIR expression had a significantly poorer prognosis with regard to overall survival and a significantly larger tumor size than those with low HOTAIR expression (p<0.01) (Fig. 1B). Clinicopathological factors were analyzed between groups (Table I), and no significant differences in other clinicopathological factors or in recurrence-free survival were noted between the high and low expression groups (data not shown).
Subsequently, we examined whether HOTAIR overexpression in HepG2 cells induced more rapid cell growth using an MTT assay. While we observed no significant differences between normal HepG2 cells (control) and HepG2 cells overexpressing HOTAIR, there was a tendency for the HOTAIR-expressing HepG2 cells to grow more rapidly than the control HepG2 cells (p=0.10) (Fig. 2).
We next investigated the relationship between HOTAIR expression and gene signatures after HOTAIR-induced H3K27me3 and EZH2 occupancy using GSEA. Based on the results of GSEA, there were no significant correlations between HOTAIR expression levels in HCC and the expression levels of signatures induced by H3K27me3 and EZH2 occupancy (Fig. 3). These results suggest that HOTAIR expression in HCC may not induce genome-wide retargeting of PRC2, unlike that noted in breast and colorectal cancers.
Discussion
In the present study, we found that high HOTAIR expression in HCC primary tumors was associated with a poor prognosis. As for clinicopathological factors, there was a significant association between HOTAIR expression and tumor size. Additionally, we found that introduction of human HOTAIR in liver cancer cells resulted in a tendency for more rapid proliferation, compared with the control cells. We speculate that HOTAIR may promote cell growth, explaining the significant association between HOTAIR expression and tumor size in HCC, and leading to poorer prognoses in HCC patients with high HOTAIR expression.
Recently, researchers have been searching for novel biomarkers to predict tumor recurrence in patients who have undergone liver transplantation (16). In the present study, we showed that high HOTAIR expression in primary HCC could be a useful marker for predicting HCC recurrence since HCC cases can be clearly categorized into high and low expression groups.
In previous studies, including ours, HOTAIR expression was shown to interact with multiple genes in cooperation with PRC2 (5), suggesting an important role for HOTAIR in tumor growth and tumorigenicity of breast and colorectal cancers. In addition, Yang et al reported that long ncRNAs, including HOTAIR, are expressed at high levels in HCC tumors in vivo(15). HOTAIR and other long ncRNAs, such as XIST, and other unidentified ncRNAs, may play significant roles in tumor growth in cooperation with PRC2 and histone modification genes in HCC (17–19). In the present study, however, GSEA results showed that the expression profiles generally induced by H3K27me3 and EZH2, a PRC2 element, were not significantly associated with HOTAIR expression in HCC. Therefore, this ncRNA may cooperate with other molecules or in different pathways in HCC, rather than functional with PRC2 complex proteins, and further research is required to fully elucidate these mechanisms.
In conclusion, certain HCC patients express the long ncRNA HOTAIR, and HCC patients with high HOTAIR expression exhibits poorer prognoses than those with low HOTAIR expression. HOTAIR expression in primary HCC tumors may be a prognostic marker in HCC; however, the clinical significance of HOTAIR expression in HCC appears to be less important than that in breast and colorectal cancers, including parameters such as histological grade, tumor depth and lymph node metastasis. Our data demonstrate the need for further study to support the use of ncRNAs as new clinical indicators of poor prognosis in HCC.
Acknowledgements
The authors thank T. Shimooka, and M. Kasagi for their technical assistance and H. Miyoshi (RIKEN BioResource Center) for providing the lentiviral vector plasmid DNA. This study was supported in part by the following grants and foundations: CREST, Japan Science and Technology Agency (JST); Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (grant nos. 20390360, 20591547, 20790960, 21591644, 21791295, 21791297, 215921014 and 21679006); the Funding Program for Next Generation World-Leading Researchers (LS094); NEDO (New Energy and Industrial Technology Development Organization) Technological Development for Chromosome Analysis; and Grant-in-Aid from the Tokyo Biochemical Research Foundation.
References
|
Ponting CP, Oliver PL and Reik W: Evolution and functions of long noncoding RNAs. Cell. 136:629–641. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Rinn JL, Kertesz M, Wang JK, et al: Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Khalil AM, Guttman M, Huarte M, et al: Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009. View Article : Google Scholar | |
|
Gupta RA, Shah N, Wang KC, et al: Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Kogo R, Shimamura T, Mimori K, et al: Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Kudo M, Izumi N, Kokudo N, et al: Management of hepatocellular carcinoma in Japan: Consensus-Based Clinical Practice Guidelines proposed by the Japan Society of Hepatology (JSH) 2010 updated version. Dig Dis. 29:339–364. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Umemura T, Ichijo T, Yoshizawa K, Tanaka E and Kiyosawa K: Epidemiology of hepatocellular carcinoma in Japan. J Gastroenterol. 44(Suppl 19): 102–107. 2009. View Article : Google Scholar | |
|
Ammerpohl O, Pratschke J, Schafmayer C, et al: Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 130:1319–1328. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
McKnight R, Nassar A, Cohen C and Siddiqui MT: Arginase-1: a novel immunohistochemical marker of hepatocellular differentiation in fine needle aspiration cytology. Cancer Cytopathol. 120:223–229. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Pan K, Liang XT, Zhang HK, et al: Characterization of BIN1 as a potential tumor suppressor and prognostic marker in hepatocellular carcinoma. Mol Med. 18:507–518. 2012.PubMed/NCBI | |
|
Andrisani OM, Studach L and Merle P: Gene signatures in hepatocellular carcinoma (HCC). Semin Cancer Biol. 21:4–9. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Qiu J, Huang P, Liu Q, et al: Identification of MACC1 as a novel prognostic marker in hepatocellular carcinoma. J Transl Med. 9:1662011. View Article : Google Scholar : PubMed/NCBI | |
|
Inoue H, Mori M, Honda M, et al: The expression of tumor-rejection antigen ‘MAGE’ genes in human gastric carcinoma. Gastroenterology. 109:1522–1525. 1995. | |
|
Subramanian A, Tamayo P, Mootha VK, et al: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Yang F, Zhang L, Huo XS, et al: Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Z, Zhou L, Wu LM, et al: Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 18:1243–1250. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Huarte M, Guttman M, Feldser D, et al: A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 142:409–419. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Braconi C, Valeri N, Kogure T, et al: Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Tsai MC, Manor O, Wan Y, et al: Long noncoding RNA as modular scaffold of histone modification complexes. Science. 329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI |