lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer
- Authors:
- Published online on: September 22, 2016 https://doi.org/10.3892/or.2016.5121
- Pages: 2960-2966
Abstract
Introduction
Esophageal cancer (EC) is one of the most common gastrointestinal cancers, which leads to the sixth ranking of cancer-related death (1,2). The treatments of EC include surgery, chemotherapy and radiation therapy. Even though the treatments of EC have made great progress, the prognosis of patients is still poor (3). That most of the patients are diagnosed at the advanced stage of EC when they are hospitalized is an important reason for high mortality (4,5). Therefore, it is very important to identify the novel molecules closely associated with the occurrence and development of EC, which will be helpful for early diagnosis and developing new target therapy to overcome the disease.
Long non-coding RNAs (lncRNAs) are a class of transcripts larger than 200 nt in length without protein coding potential. lncRNAs were initially thought to be 'transcriptional noise' resulting from low RNA polymerase fidelity (6). However, emerging evidence have demonstrated that lncRNAs play pivotal roles in many biological processes, such as regulation of gene expression, chromatin remodeling, X chromosome inactivation, post-transcriptional modification and translational control (7–10). Dysregulation of lncRNAs have been found in various diseases including cancers (11,12). Many studies have reported that the abnormally expressed lncRNAs have multiple functions in regulation of malignant phenotypes, such as proliferation, apoptosis, cell cycle, metabolism or cell migration (13–15). Until now, only a few functional lncRNAs in EC have been identified. It is not fully clear whether other lncRNAs are involved in carcinogenesis and development of EC.
lncRNA human urothelial carcinoma associated 1 (UCA1) gene is located in chromosome 19p13. It has been reported that UCA1 is significantly upregulated in many types of cancer, such as hepatocellular carcinoma, breast, colorectal and gastric cancer, suggesting that UCA1 may function as an important oncogene in human cancers (16–18). UCA1 overexpression promotes cancer progression by regulating mTOR, Wnt or Akt signaling pathway (19–21). However, the role of UCA1 in EC and its underlying mechanism remains unclear.
In the present study, we demonstrated that UCA1 was significantly upregulated in EC tissues and associated with poor prognosis. Overexpression of UCA1 promoted the proliferation of EC cells, while silence of UCA1 inhibited EC cell growth. Furthermore, we found that Sox4 was a direct target gene of UCA1. UCA1 regulated Sox4 expression through functioning as a competing endogenous RNA (ceRNA). UCA1 completed with Sox4 mRNA for miR-204, a key tumor suppressive miRNA in various cancers. The present study provides the first evidence that UCA1 promotes cell proliferation through Sox4 in EC, suggesting that UCA1 and Sox4 may be potential therapeutic targets for EC.
Materials and methods
Patients
A total of EC tissues and paired adjacent normal esophagus tissues from 66 patients with EC diagnosed by histopathological examination and undergoing resection between 2009 and 2015 were collected. Clinical characteristics of these patients were collected retrospectively for all patients. Informed consent was obtained from all the patients.
Cell culture
EC9706 and KYSE cell lines were obtained from the Chinese Academy of Sciences Cell Bank. Cells were cultured in RPMI-1640 (HyClone Laboratories, Inc., Logan, UT, USA) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U/ml penicillin and 100 U/ml streptomycin. These cell lines were cultured in humidified 5% CO2 at 37°C.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using TRIZol reagent (Invitrogen, Carlsbad, CA, USA). First strand cDNA was generated using the Reverse EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgene Biotek, Ltd., Hyderabad, India). qRT-PCR was performed by using SYBR Green Master Mix (Roche) reagent in ABI 7500 real-time PCR instrument. ACTB was employed as an endogenous control. The relative levels of gene expression were calculated by the 2−ΔΔCt method. Primers used for RT-RCR are as follows: UCA1-F, 5′-GAG GATTCCCAGCCATATGAAG-3′ and UCA1-R, 5′-CGGCAGTTGGTGTGCTATAA-3′; Sox4-F, 5′-CCAACTCCTTAGTGCCGATT-3′ and Sox4-R, 5′-TCTCTAGCACTTAGTCCCTCTC-3′; ACTB-F, 5′-CACCAGGCACCCAGTTTAAT-3′ and ACTB-R, 5′-AGTCTCTGCTCTCTCTTCCTATG-3′.
Cell proliferation assay
Cell proliferation was determined by the Cell Counting kit-8 (CCK-8) assay. Cells (2×103) were seeded in 96-well plates. Each well was incubated with WST-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) for 3 h, and the absorbance was measured at 450 nm using a spectrophotometer. All experiments were performed in triplicate.
Western blotting
Western blotting was performed with the standard protocol. Primary antibodies used were rabbit anti-Sox4 (Abcam) and mouse anti-β-actin (Proteintech, Chicago IL, USA).
Construction of stable UCA1 overexpressed or knockdown cells
To construct UCA1 knockdown cells, cells were infected with lentivirus particles expressing shRNA against UCA1 and selected by puromycin (1 μg/ml) for one week. The target sequence is as follow: GCCATATGAAGACACCCTA. To construct stable UCA1 overexpressed cells, cells were transfected with pcDNA3.1 containing full-length UCA1 and selected with G418 (400 μg/ml) for one week.
siRNA transfection
siRNAs against Sox4 were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The target sequence against Sox4 is as follows: GGACTAAGTGCTAGAGACT.
Luciferase reporter assay
The Sox4 3′ untranslated region (3′UTR) was cloned to luciferase reporter plasmid, pmirGLO. The pmirGLO-Sox4 was cotransfected with miR-NC (negative control) or miR-204 mimics into cells using TurboFect (Thermo Fisher Scientific). The relative luciferase activity was normalized to Renilla luciferase activity 48 h after transfection. The luciferase activity was measured using a Dual-luciferase reporter gene assay system (Promega, Madison, WI, USA).
RNA immunoprecipitation (RIP)
Anti-MS2 RIP and anti-AGO2 RIP was performed as previously describe by using the EZ-Magna RNA immunoprecipitation kit (Millipore) (9).
Statistical analyses
All statistical analyses were performed by SPSS 20.0 software. For comparisons, Student's t-test and Pearson's Chi-square test were performed as appropriate. A value of P<0.05 was considered to indicate a statistically significant result.
Results
The expression of UCA1 is significantly upregulated in human EC tissues and associated with poor prognosis
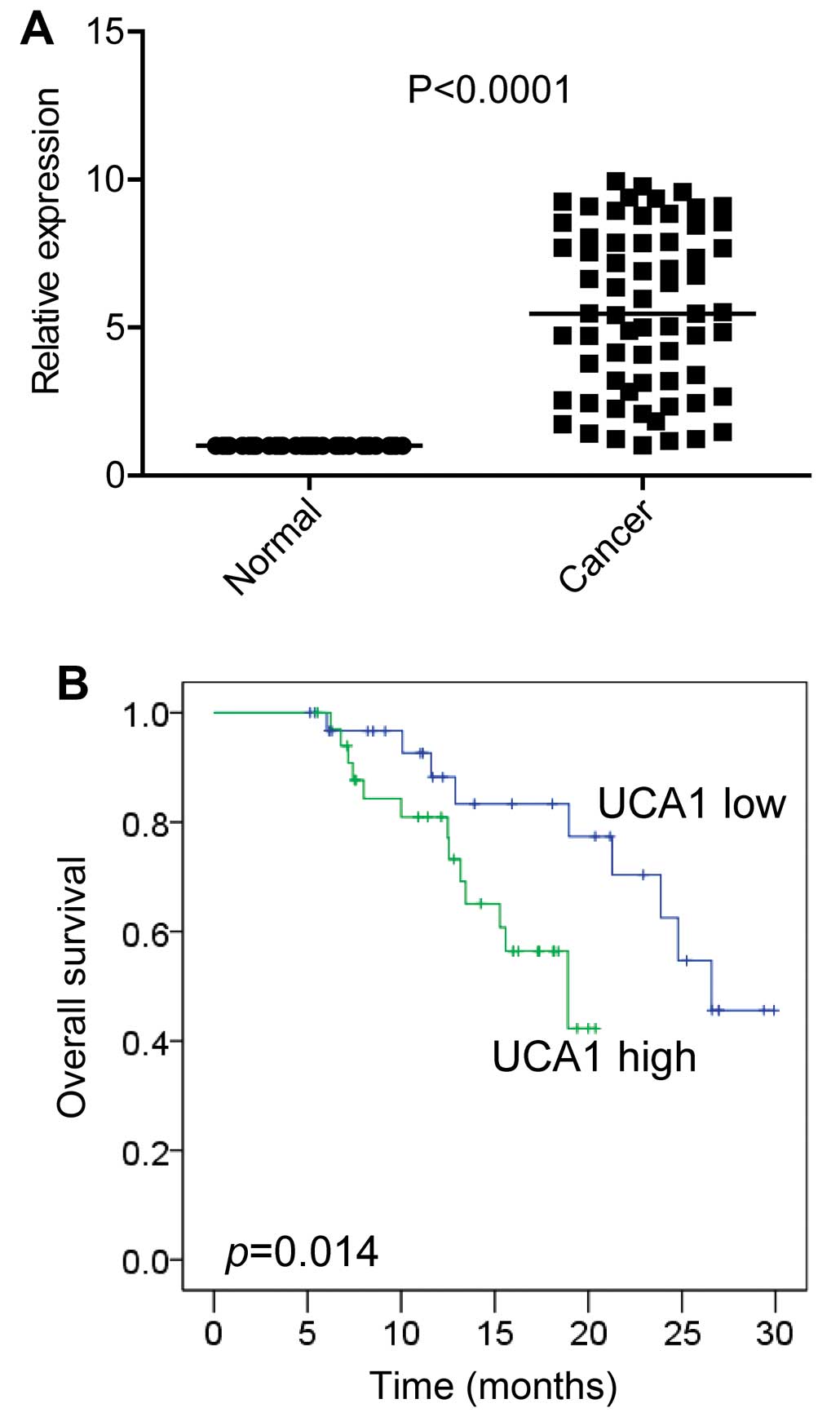
To determine the role of UCA1 in EC, the UCA1 expressions were detected in EC tissues and their paired adjacent normal tissues from 66 EC patients by using qRT-PCR. We found that UCA1 expression was significantly increased in EC tissues compared with the paired normal tissues (Fig. 1A).
To gain insight to the clinical significance of UCA1 expression, we analyses the correlation of UCA1 expression with clinicopathological features (Table I). These 66 patients were classified into two groups using the median expression level of UCAL1 in EC tissues as the cut-off value. Notably, higher UCA1 expression in EC was significantly associated with poor differentiation and higher TNM stage.
Table IThe correlation between UCA1 expression and clinicopathological characteristics in 66 patients with EC. |
Next, we determined the association of UCA1 and prognosis by performing Kaplan-Meier analysis. The results showed that patients with higher UCA1 expression had a significantly poorer prognosis compared to patients with lower UCA1 expression (Fig. 1B).
UCA1 promotes EC cell proliferation
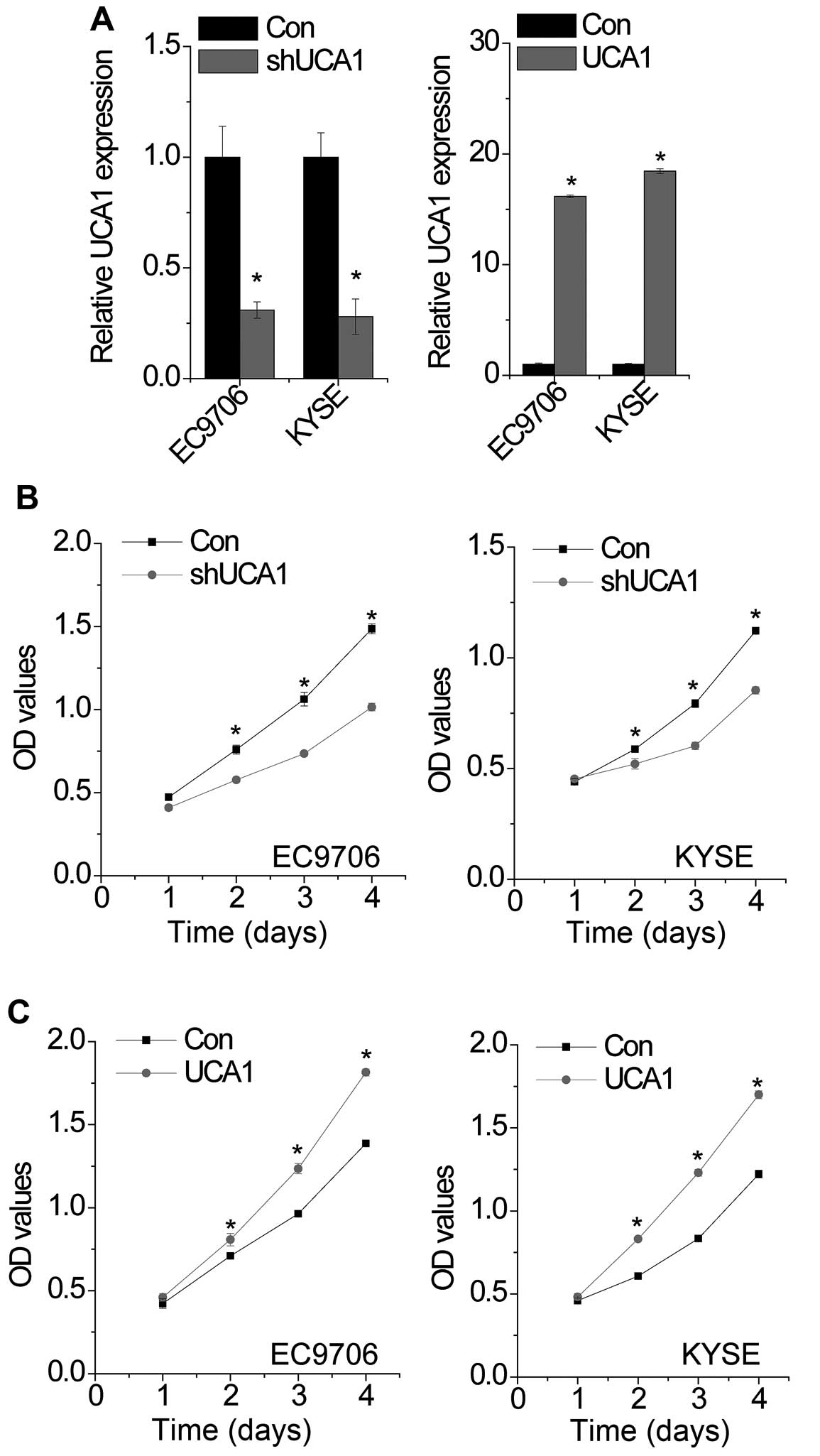
To determine the function of UCA1 in EC cells, we constructed stable cells with knockdown or overexpression of UCA1 (Fig. 2A). We then detected the effect of UCA1 on proliferation by CCK-8 assay. The results showed that both UCA1 overexpressed EC9706 and KYSE cells was significantly accelerated compared with that of the control cells (Fig. 2B), while knockdown of UCA1 effectively inhibits cell proliferation (Fig. 2C).
UCA1 is physically associated with miR-204
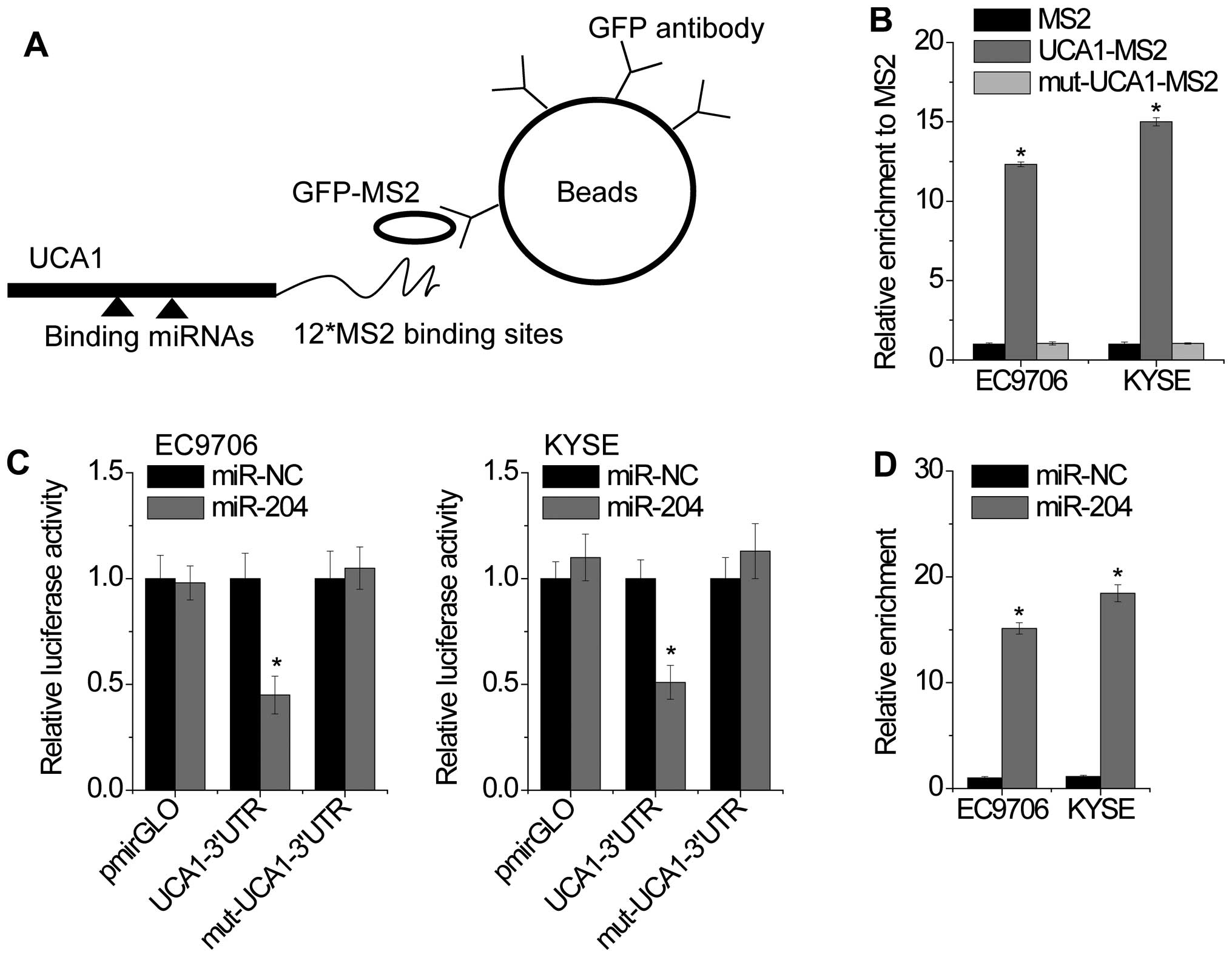
It has been reported that lncRNAs can function as microRNA (miRNA) sponge to regulate the activity of miRNAs (22). A recent study reported that UCA1 could sponge endogenous miR-204 and inhibit its activity in colorectal cancer (16). However, the direct interaction of UCA1 and miR-204 in EC remains unclear. To confirm the direct binding between UCA1 and miR-204, we performed RIP assay to pull down miRNAs interacted with UCA1. The results showed that UCA1 was significantly enriched for miR-204 compared to the empty vector (MS2) and UCA1 with mutations in miR-204 targeting sites (mut-UCA1-MS2) (Fig. 3A and B). In addition, we constructed luciferase reporter plasmids containing the wild-type UCA1 or mutant UCA1 (mut-UCA1) with mutated miR-204 binding sites. We found that miR-204 overexpression decreased the luciferase activities of the wild-type UCA1 reporter vector but not empty vector or mutant UCA1 reporter vector (Fig. 3C). Finally, we performed anti-AGO2 RIP to detect whether UCA1 was regulated by miR-204 in an AGO2-dependent manner (Fig. 3D). Endogenous UCA1 pull-down by AGO2 was significantly enriched in miR-204 overexpressed cells, suggesting that miR-204 is a bona fide UCA1-targeting miRNA.
UCA1 functions as a ceRNA of Sox4
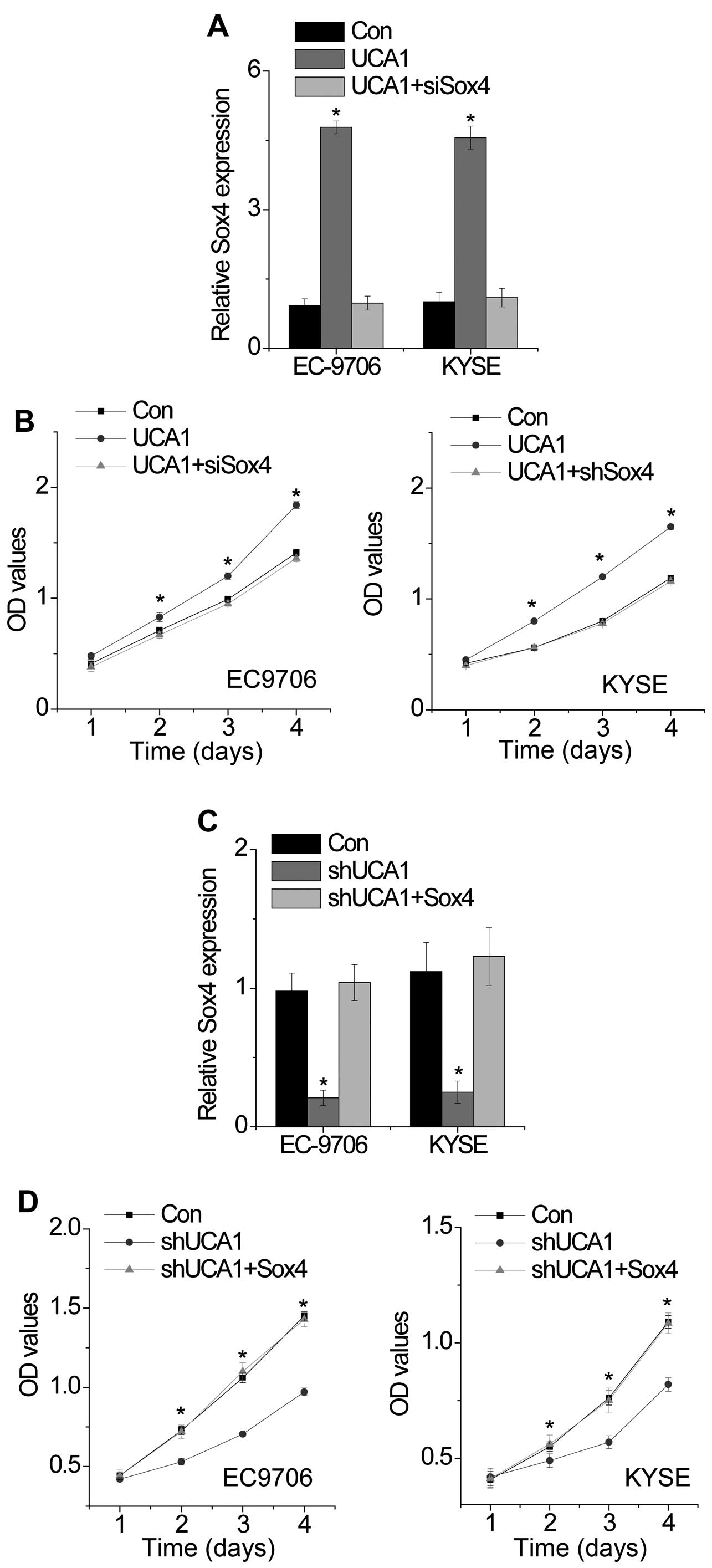
Recent studies have reported that Sox4 is a target gene of miR-204 (23,24). We suspected that UCA1 may regulate Sox4 expression through functioning as a ceRNA. To confirm this hypothesis, we detected the mRNA and protein level of Sox4 in UCA1 overexpressed and UCA1 knockdown cells. Surprisingly, we found that overexpression of UCA1 increased Sox4 expression, while silence of UCA1 suppressed Sox4 expression (Fig. 4A and B). For rescue experiment, we transiently transfected wild-type UCA1 or mutant UCA1 overexpressed cells with miR-204. Overexpression of wild-type UCA1, but not the mutant UCA1, increased Sox4 expression (Fig. 4C). Overexpression of miR-204 abolished the increase of Sox4 induced by UCA1. In addition, we suppressed miR-204 expression in UCA1 knockdown cells and found that inhibition of miR-204 rescued the decrease of Sox4 induced by UCA1 knockdown (Fig. 4D).
To further confirm whether the UCA1 influenced Sox4 expression depends on the regulation of Sox4 3′UTR, we constructed luciferase reporters containing Sox4 3′UTR (pmirGLO-Sox4). The luciferase plasmid (pmirGLO-Sox4) was transfected into the wild-type or mutant UCA1 overexpressed cells with or without miR-204. We found that overexpression of wild-type UCA1, but not the mutant UCA1, increased the luciferase activity of Sox4 3′UTR, while miR-204 abolished this effect (Fig. 4E). Similarly, the silence of UCA1 decreased the luciferase activity of Sox4 3′UTR, which were rescued by inhibition of miR-204 (Fig. 4F).
We next detected the expression levels of Sox4 in 66 EC and paired normal tissues. As shown in Fig. 4G, UCA1 expression was significantly correlated with Sox4 mRNA expression. All these results suggest an important role of UCA1 in regulating Sox4 expression by competitively binding miR-204.
UCA1 promotes cell proliferation through regulation of Sox4
Finally, we detected whether UCA1 promotes cell proliferation through regulation with Sox4 expression. We transfected siRNA against Sox4 in UCA1 overexpressed cells (Fig. 5A). We found that silence of Sox4 abolished the cell proliferation induced by UCA1 overexpression (Fig. 5B). Consistently, overexpression of Sox4 rescued the inhibition of cell proliferation mediated by UCA1 knockdown (Fig. 5C and D). These results suggest that UCA1 promotes cell proliferation through regulation of Sox4.
Discussion
The present study demonstrated that UCA1 was significantly upregulated in EC tissues compared with paired adjacent normal esophagus tissues. The UCA1 expression is closely correlated with TNM stage and tumor differentiation. In addition, the high-level expression of UCA1 predicted poor prognosis. The results of this study showed the pathological roles of UCA1 in promoting cell proliferation. Therefore, this study indicated that UCA1 functions as an oncogene in EC and can be a potential biological target of the therapy for EC.
Increasing evidence have demonstrated that lncRNAs play important roles in EC. For example, lncRNA POU6F2-AS2 is involved in the DNA damage response and regulates cell survival after ionizing radiation through interacting with Ybx1 protein and regulating its chromatin localization (25). linc-POU3F3 is significantly increased in EC and contributes to the development of ESCC by interacting with EZH2 to promote methylation of POU3F3 (26). However, to the best of our knowledge, the mechanism related to ceRNA in EC remains poorly understood. In the present study, we found that UCA1 promotes cell proliferation through functioning as a ceRNA of Sox4. UCA1 completed with Sox4 for common miR-204 which is an important tumor suppressor miRNA in various cancers (27–29). UCA1 directly interacted with miR-204 and decreased the binding of miR-204 to Sox4 3′UTR, which suppressed the degradation of Sox4 mRNA by miR-204. A previous study also showed the interaction of UCA1 and miR-204 could enhance 5-fluorouracil resistance in colorectal cancer (16). UCA1 could also function as ceRNAs of other mRNAs in other cancers, such as MMP14, ERBB4 and ZEB1/2 (30–32). The present study enriched the ceRNA network regulated by UCA1.
Sox4, the member of the Sox (Sry-related high mobility group box) family of transcription factors, is as a master mediator in tumorigenicity and cancer stemness (33). Sox4 is upregulated in various cancers, such as colorectal, prostate cancer and EC (34–36). The expression of Sox4 was post-transcriptionally regulated by miR-363, miR-204 and miR-187 (23,37,38). However, whether Sox4 expression is regulated by lncRNA remains unclear. The present study, for the first time, demonstrated that Sox4 was regulated by lncRNA UCA1 via a ceRNA mechanism. We found that overexpression of UCA1 increased Sox4 expression, while silence of UCA1 suppressed Sox4 expression. Furthermore, UCA1 expression was positively correlated with Sox4 expression in 66 EC tissues. These results strongly suggest that Sox4 is a direct target gene of UCA1.
In summary, the present study demonstrated that UCA1 as an oncogene promotes cell proliferation in a ceRNA manner in EC. As a direct target of UCA1, miR-204-Sox4 mediated the role of UCA1 in cell proliferation. These findings indicate that UCA1 is an important molecular for tumor growth and could be an effective target for EC therapy.
References
|
Sun X, Chen W, Chen Z, Wen D, Zhao D and He Y: Population-based case-control study on risk factors for esophageal cancer in five high-risk areas in China. Asian Pac J Cancer Prev. 11:1631–1636. 2010. | |
|
Hongo M, Nagasaki Y and Shoji T: Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 24:729–735. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Song Q, Liu H, Wang J, Jia Y, Liu Y, Wang N, Tan B, Guan S, An D and Cheng Y: Dinner-to-bed time and post-dinner walk: New potential independent factors in esophageal cancer development. J Cancer Res Clin Oncol. 140:817–821. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Jemal A, Center MM, DeSantis C and Ward EM: Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Tang WR, Fang JY, Wu KS, Shi XJ, Luo JY and Lin K: Epidemiological characteristics and prediction of esophageal cancer mortality in China from 1991 to 2012. Asian Pac J Cancer Prev. 15:6929–6934. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Louro R, Smirnova AS and Verjovski-Almeida S: Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar | |
|
Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM and Morrisey EE: Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 28:1363–1379. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al: A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 26:344–357. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Rühle F and Stoll M: Long non-coding RNA databases in cardiovascular research. Genomics Proteomics Bioinformatics. 14:191–199. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T and Li X: Competing endogenous RNA networks in human cancer: Hypothesis, validation, and perspectives. Oncotarget. 7:13479–13490. 2016.PubMed/NCBI | |
|
Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, et al: LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 18:431–442. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al: Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. Apr 11–2016. View Article : Google Scholar | |
|
Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia M, Zhan Y, Lin J, Chen Z, He A, et al: Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 376:155–164. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204–5p. Sci Rep. 6:238922016. View Article : Google Scholar | |
|
Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ, Gong PH, Lu S, Zhao ZJ, He ML, et al: HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 6:235212016. View Article : Google Scholar : PubMed/NCBI | |
|
Gu W, Gao T, Sun Y, Zheng X, Wang J, Ma J, Hu X, Li J and Hu M: LncRNA expression profile reveals the potential role of lncRNAs in gastric carcinogenesis. Cancer Biomark. 15:249–258. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Wu W, Zhang S, Li X, Xue M, Cao S and Chen W: Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One. 8:e739202013. View Article : Google Scholar : PubMed/NCBI | |
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Li Z, Li X, Wu S, Xue M and Chen W: Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 105:951–955. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, et al: Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 7:109822016. View Article : Google Scholar : PubMed/NCBI | |
|
Yu CC, Chen PN, Peng CY, Yu CH and Chou MY: Suppression of miR-204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget. 7:20180–20192. 2016.PubMed/NCBI | |
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 73:990–999. 2013. View Article : Google Scholar : | |
|
Liu J and Sun X, Zhu H, Qin Q, Yang X and Sun X: Long noncoding RNA POU6F2-AS2 is associated with esophageal squamous cell carcinoma. J Biochem. Mar 31–2016.(Epub ahead of print) pii: mvw025. | |
|
Li W, Zheng J, Deng J, You Y, Wu H, Li N, Lu J and Zhou Y: Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 146:1714–1726.e5. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Z, Long J, Du R, Ge C, Guo K and Xu Y: miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 37:8327–8335. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wu ZY, Wang SM, Chen ZH, Huv SX, Huang K, Huang BJ, Du JL, Huang CM, Peng L, Jian ZX, et al: MiR-204 regulates HMGA2 expression and inhibits cell proliferation in human thyroid cancer. Cancer Biomark. 15:535–542. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Ding M, Lin B, Li T, Liu Y, Li Y, Zhou X, Miao M, Gu J, Pan H, Yang F, et al: A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget. 6:7686–7700. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Y, Jiang Y, Wan Y, Zhang L, Qiu J, Zhou S and Cheng W: UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. Feb 11–2016.Epub ahead of print. | |
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 371:99–106. 2016. View Article : Google Scholar | |
|
Xue M, Pang H, Li X, Li H, Pan J and Chen W: Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016. View Article : Google Scholar | |
|
Ye X and Weinberg RA: Epithelial-mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 25:675–686. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Bilir B, Osunkoya AO, Wiles WG IV, Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD and Moreno CS: SOX4 is essential for prostate tumorigenesis initiated by PTEN ablation. Cancer Res. 76:1112–1121. 2016. View Article : Google Scholar | |
|
Sun R, Jiang B, Qi H, Zhang X, Yang J, Duan J, Li Y and Li G: SOX4 contributes to the progression of cervical cancer and the resistance to the chemotherapeutic drug through ABCG2. Cell Death Dis. 6:e19902015. View Article : Google Scholar : PubMed/NCBI | |
|
Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA and Andl CD: SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 14:242015. View Article : Google Scholar : PubMed/NCBI | |
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and Li X: MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, Shi J, Sun X, Liu Y, Ding Y, et al: MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 373:203–213. 2016. View Article : Google Scholar : PubMed/NCBI |