Multidrug resistance protein 5 affects cell proliferation, migration and gemcitabine sensitivity in pancreatic cancer MIA Paca‑2 and PANC‑1 cells
- Authors:
- Published online on: November 16, 2023 https://doi.org/10.3892/or.2023.8666
- Article Number: 7
-
Copyright: © He et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Pancreatic cancer is one of the leading causes of cancer-related death worldwide; it is an aggressive disease that is usually advanced at the time of diagnosis. Despite all of the developments in pancreatic cancer research, the death to incidence ratio has not significantly improved in the last couple of decades (1–3). Pancreatic malignancy has a 5-year survival rate of 8% for all stages (4). The disease advances asymptomatically in 80% of patients and is generally discovered in the later stages, by which time the only treatment option is chemotherapy, and its surgical removal is not possible (5). Even after chemotherapy, the chances of survival are not significantly high due to multidrug resistance (MDR), which impairs the efficacy of the combination chemotherapy (5). Various genetic mutations associated with pancreatic ductal adenocarcinoma (PDAC) and dysregulation of several signalling pathways make PDAC highly heterogeneous (6). Since 1997 to up until now, gemcitabine (a cyclic nucleotide) has been used as the first-line treatment drug for PDAC (6). While gemcitabine has marginally improved the patients' survival rate, the overall survival of patients with advanced disease remains below six months. Due to the low efficacy of gemcitabine, efforts are being made to increase the survival of the patients (6). MDR is an intricate process and there are a number of factors working in parallel, which may influence the process, e.g., insufficient drug delivery due to a high level of fibrosis and low vascularity, accelerated drug metabolism and DNA repair, blocking of apoptotic pathways, metabolic changes and the presence of highly resistant stem-like cells (7–10). Although accumulating preclinical evidence indicates that ATP-binding cassette (ABC) transporter-mediated drug efflux contributes to MDR (11,12), the lack of success of ABC transporter inhibitor in clinical trials suggests our knowledge in this area is still limited or biased (13).
ABC transporters belong to a highly conserved family of proteins, and to date, 48 genes and one pseudogene have been identified in humans (12,14–16). The family is further subdivided into seven subfamilies (ABCA-G) based on the premise of their structure and sequence (16–18). ABC transporters are transmembrane proteins, and as the name suggests, ABC transporters utilise energy from ATP hydrolysis for the efflux of their substrates (4,17,19). Overexpression of one of the ABC transporter subfamilies was found in PDAC (20–22). The existence of ABC transporter expression signatures in PDAC and their association with clinicopathological characteristics of the tumours has been studied by Mohelnikova-Duchonova et al (23) and dysregulation of the expression of several members of the ABC family has been observed. Upregulation of ABCB4, ABCB11, ABCC1, ABCC3, ABCC5 [aka., multidrug resistance protein 5 (MRP5)], ABCC10 and ABCG2 has been noted in PDAC, compared to nonneoplastic tissues (4–6,24).
MRPs are integral membrane proteins mediating the ATP-dependent export of organic anions out of cells. Previously, it has been observed that MRP4 and MRP5 were localised in duct cells, acinar cells and pancreatic cancer cells (22,25). The MRP5 mRNA level was significantly higher in pancreatic carcinoma tissue compared to normal pancreatic tissue (6,11,17,26). The MRP5 A-2G AA genotype also showed a significant association with overall survival in patients with pancreatic cancer (27). MRP5 has also been demonstrated to confer gemcitabine resistance in various in vitro cell culture models (5,6,17,26,28,29).
Apart from their drug efflux abilities, emerging evidence suggests the contributions of ABC transporters to cancer biology in either a substrate efflux-dependent or -independent manner (12,30–32), leading to cancer cell proliferation, differentiation, invasion and/or migration. Thus, gaining better insight into the role of ABC transporters in both cancer biology and tumour resistance may lead to new and better anti-cancer strategies. In the present study, it was hypothesised that MRP5 transporter possibly has pleotropic roles in cancer development, in addition to its gemcitabine efflux actions in pancreatic cancer cells (16). The role of MRP5 in cancer hallmarks of pancreatic cancer cells was investigated by using lentiviral short-hairpin (sh)RNA gene modulation techniques. Stable knockdown of the MRP5 gene was achieved and its effects on MRP5 expression, gemcitabine cytotoxicity and growth and migration of pancreatic cancer cells were measured.
Materials and methods
Cell culture
The human pancreatic cancer cell lines MIA Paca-2 and PANC-1 were obtained from the American Type Culture Collection. MIA Paca-2 and PANC-1 cell lines were cultured using complete DMEM (cat. no. 11965-092; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) FBS (cat. no. MG-FBS0820; MediRay) and 2 mmol/l L-glutamine (cat. no. 20530081; Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5% carbon dioxide at 37°C.
Lentiviral transduction
Cells were seeded at a density of 1.2×104 cells per well in a 96-well plate and incubated for 24 h. The culture medium was then replaced with 110 µl fresh medium containing hexadimethrine bromide (final concentration, 8 µg/ml) (cat. no. H9268; Sigma-Aldrich; Merck KGaA). An aliquot of 1 µl custom-designed shRNA lentiviral particles (Sigma-Aldrich; Merck KGaA) targeting the MRP5 gene was added to MIA Paca-2 and PANC-1 cells (multiplicity of infection, 2) and incubated for 20 h. MISSION® Non-mammalian shRNA control transduction particles (cat. no. SHC002V; Sigma-Aldrich; Merck KGaA) were used as a negative control. After a 20-h incubation, the pooled population was transferred to two 75 mm Petri Dishes and the transduced cells were selected in medium containing 0.4 µg/ml puromycin (cat. no. A1113803; Thermo Fisher Scientific, Inc.). For further investigation, two clones of each cell type were randomly picked up, which were MIA Paca-2 clones 1 (M c1) and 2 (M c2) and PANC-1 clones 1 (P c1) and 2 (P c2). The shRNA target sequence is 5′-CCACATCTTCAATAGTGCTAT-3′. The sequence is unique in the human transcriptome, which only targets MRP5 mRNA and its variants. The insert sequence of the scrambled control is 5′-CCGGCAACAAGATGAAGAGCACCAACTC-GAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
RNA extraction from cultured cells was carried out using TRIzol® reagent (cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The total RNA was quantified using a NanoDrop® ND-1000 UV spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc.). Primer sequences as listed in Table I (33,34) and were purchased from Integrated DNA Technologies, Inc. RT-qPCR amplification and analysis were performed with a Light Cycler 480 Instrument (Roche Diagnostics) using the LightCycler® EvoScript RNA SYBR® Gren I Master one-step RT-qPCR kit (cat. no. 07800134001; Roche Applied Science). For each reaction, an aliquot of 5 µl RNA sample (10 ng) was mixed with 1 µl primer mix (final concentration 0.4 µM), 4 µl master mix (5× conc.) and 10 µl PCR grade water. The reaction conditions of RT-qPCR were 60°C for 15 min (reverse transcription) and 95°C for 10 min (enzyme activation), followed by 45 cycles of 95°C for 10 sec (denaturation) and 59°C for 30 sec (annealing and extension). MRP5 mRNA expression in each sample was normalised to the reference gene GAPDH and the results were analysed using the comparative threshold cycle method (35).
MRP5 surface staining
MRP5 expression was confirmed by surface immunostaining of the protein. Cells were harvested and aliquoted at 1×106 cells into Eppendorf tubes. After fixation in 1% paraformaldehyde (PFA) (cat. no. P6148; Sigma-Aldrich; Merck KGaA) at 4°C for 15 min, the cells underwent permeabilization in 0.2% saponin (cat. no. 47036-50G-F, Sigma-Aldrich; Merck KGaA) at 4°C for 15 min, blocking in 5% bovine serum albumin (BSA) (cat. no. NZ21-69100-038; pH Scientific) at room temperature for 15 min, and staining with anti-MRP5 rat monoclonal antibody (M5II-54) (1:20 in 2% BSA; cat. no. MA1-35684; Thermo Fisher Scientific, Inc.) or Rat IgG2α Isotype Control primary antibody (1:20 in 2% BSA; cat. no. 02-9688, Thermo Fisher Scientific, Inc.) at 4°C for 1 h. The cells were then stained with goat anti-Rat IgG H&L (Alexa Fluor® 488) secondary antibody (1:200 in 2% BSA; cat. no. ab96887; Abcam) at 4°C for 1 h. The cells were washed three times with PBS containing 0.1% NaN3 and 0.1% saponin between each step. After secondary antibody incubation, the cells were then washed and resuspended in 500 µl 1% PFA. Fluorescence was detected at 488 nm excitation and 525 nm emission wavelength using a flow cytometer (MoFlo XDP; Beckman Coulter, Inc.). The mean fluorescence intensity was determined using Kaluza Flow cytometry software version 2.2.1 (Beckman Coulter, Inc.).
MRP5 substrate accumulation
Steady-state cellular accumulation of an MRP5 substrate, 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM) (cat. no. B8806; Thermo Fisher Scientific, Inc.) was determined using a flow cytometer (MoFlo XDP; Beckman Coulter, Inc.). An MRP5 inhibitor, benzbromarone (cat. no. B5774; Sigma-Aldrich; Merck KGaA), was used as a positive control to validate the function of MRP5. MIA Paca-2 and PANC-1 cells were washed with FBS- and phenol red-free DMEM (cat. no. 31053028; Thermo Fisher Scientific, Inc.) and were resuspended in the same medium to achieve a density of 0.5×106 cells/ml. The accumulation of BCECF was performed by incubating 1 ml cell suspension with benzbromarone (50 µM) or the vehicle (0.1% DMSO) at 37°C for 30 min, followed by incubation with 0.25 µM BCECF-AM at 37°C for 15 min. The accumulation was terminated by adding ice-cold PBS. The cells were then washed with ice-cold PBS and resuspended in 500 µl 1% PFA. The samples were analysed with a standard laser for excitation at 488 nm and a bandpass filter at 525 nm to detect fluorescence. The mean fluorescence intensity was determined using Kaluza Flow cytometry software version 2.2.1 (Beckman Coulter, Inc.).
Cytotoxicity assay
The gemcitabine-induced growth inhibition was first determined in 2D-cultured pancreatic cancer cells using MTT assay. Cells were seeded at a density of 5,000 cells per well in 96-well plates and incubated for 24 h to allow cells to attach to the well surface. After incubation, cells were treated with gemcitabine (cat. no. G6423; Sigma-Aldrich; Merck KGaA) at various concentrations for 72 h. Gemcitabine stock solution at a concentration of 50 mM was prepared by dissolving gemcitabine powder in Milli-Q water. After treatment, the drug solution from each well was replaced with fresh complete medium, followed by the addition of 10 µl 12 mM MTT (cat. no. M2128; Sigma-Aldrich; Merck KGaA) stock solution. The cells were then incubated for 4 h at 37°C before adding 150 µl DMSO to each well. The cell viability was quantified by measuring the absorbance at 570 nm normalised to the mean absorbance of vehicle control. The IC50 values for gemcitabine-induced growth inhibition were determined using nonlinear regression in PRISM® software (version 8.0; GraphPad; Dotmatics).
For 3D culture models, MIA Paca-2 and PANC-1 clones were allowed to grow in 96-well low-adhesion plates (cat. no. CLS7007; Sigma-Aldrich; Merck KGaA) for 10 days, followed by 72-h incubation with different concentrations of gemcitabine. After incubation, the drug-containing medium was removed and the cells were stained with 100 µl 1X PrestoBlue (cat. no. A13261; Thermo Fisher Scientific, Inc.) at 37°C for 1 h and the fluorescence was determined at an excitation of 560 nm/emission of 590 nm using a SPARK® Microplate Spectrofluorometer (Molecular Devices, LLC).
In another experiment, the cytotoxicity of gemcitabine was assessed using a 2D colony formation assay. Cells were seeded in 96-well plates at a density of 1,000 cells per well. After 1 day, cells were treated with 2.5, 5 and 10 nM gemcitabine for 3 days, followed by incubation with drug-free medium for 7 days. The medium was changed every 3 days. After incubation, cells were fixed with 4% PFA for 15 min and stained with 0.5% crystal violet (cat. no. C6158; Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. The stained cells were washed with tap water and air-dried for a few mins before being analysed under a microscope.
Apoptosis assay
Cells were seeded at a density of 5,000 cells per well in 96-well plates. After overnight incubation at 37°C and 5% CO2, MIA Paca-2 and PANC-1 cells were treated with different concentrations of gemcitabine for 24 and 48 h, respectively. To terminate the drug exposure, the drug-containing medium was replaced with 100 µl of diluted CellEvent™ Caspase-3/7 detection reagents (final concentration 2 µM) (cat. no. C10427; Thermo Fisher Scientific, Inc.) and cells were then incubated for 60 min at 37 °C. The fluorescence was measured at an excitation of 467 nm/emission of 539 nm on a SPARK® Microplate Spectrofluorometer (Molecular Devices, LLC). Images were also acquired with a Leica DMi8 inverted fluorescence microscope (Leica Microsystems, GmbH).
Colony-formation assay
Cells were seeded in 6-well plates at a density of 200 cells per well, followed by a 10-day incubation at 37°C and 5% CO2. The medium was changed every 3 days. After incubation, cells were fixed with 4% PFA for 15 min and stained with 0.5% crystal violet for 15 min at room temperature. The stained cells were washed with tap water and air-dried for a few mins before being analysed under an inverted microscope (Zeiss AG). Colonies of >50 cells were scored for survival and counted using ImageJ 1.52g software (National Institutes of Health).
Wound-healing assay
Cells in complete DMEM were seeded into 6-well plates at a density of 3×105 cells per well and allowed to grow until a confluent cell layer was obtained. A linear scratch was made in the plate using a sterile 200 µl pipette tip (cat. no. 1030-260-000-9; Thermo Fisher Scientific, Inc.). Cells were washed and then incubated with DMEM supplemented with 0.5% FBS to prevent apoptosis (36). Images were acquired at 0, 24, 48 and 72 h for MIA Paca-2 cells and 0, 16, 24 and 40 h for PANC-1 cells with a Leica DMi8 inverted fluorescence microscope (Leica Microsystems GmbH). Cell migration was analysed using ImageJ 1.52g software.
Transwell migration assay
Cells in serum-free DMEM were seeded in the Corning® Costar® Transwell® cell culture inserts (cat. no. CLS3464; Sigma-Aldrich; Merck KGaA) at a density of 3×105 cells per insert. The inserts were placed in 12-well plates and the bottom wells were filled with 1 ml complete medium. The cells were incubated at 37°C and 5% CO2 for 48 h. After this incubation, the non-migrated cells on the upper side of the membrane were removed using a cotton swab. The migrated cells on the lower side of the membrane were fixed with 4% PFA and stained with 0.1% crystal violet in 2% ethanol. Images were taken with a Leica DMi8 inverted fluorescence microscope (Leica Microsystems GmbH) and analysed using ImageJ 1.52g software.
In silico analysis
The main software and tools used for molecular docking studies were AlphaFold (https://alphafold.ebi.ac.uk/) (37,38), PubChem (https://pubchem.ncbi.nlm.nih.gov/), Openbabel (39), AutoDock 4.2 (40), Protein-Ligand Interaction Profiler (https://plip-tool.biotec.tu-dresden.de) (41), ProteinsPlus (https://proteins.plus) (40,42) and LigPlot (43). The ligands were downloaded from PubChem in SDF format. The protein structure of MRP5 was downloaded from AlphaFold. The ligands in SDF file format were converted to pdb format using Openbabel. All of the ligands were docked with their receptor molecule using AutoDock 4.2. Results were evaluated and visualised using Protein-Ligand Interaction Profiler, ProteinsPlus and LigPlot.
Statistical analysis
Data were presented by descriptive statistics as the mean ± standard deviation. Linear and non-linear regression were carried out using PRISM® software (version 8.0; GraphPad; Dotmatics). The statistical analysis was performed by a one-way analysis of variance (ANOVA) with a Dunnett's post-hoc test or a two-way ANOVA with Sidak's post-hoc test. All data analysed using ANOVAs should meet the assumptions of equal variance and homogeneity. P<0.05 was considered to indicate statistical significance.
Results
MIA Paca-2 and PANC-1 cells endogenously express high MRP5
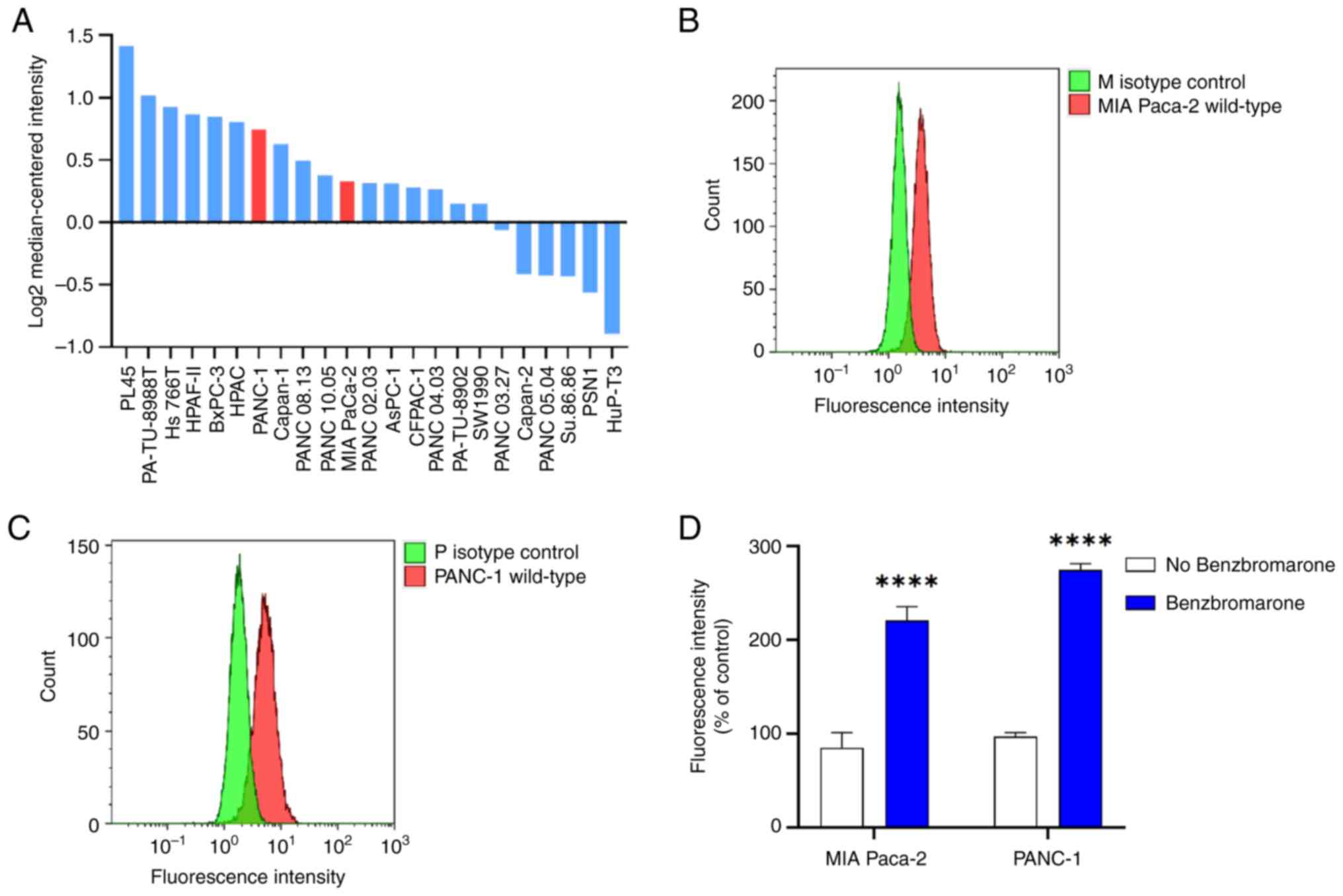
High MRP5 mRNA expression was observed in PANC-1 (ranked 7/23) and MIA PaCa-2 (ranked 11/32) cells compared to other pancreatic cancer cell lines based on the Wagner dataset stored in ONCOMINE (https://www.oncomine.org) (Fig. 1A). The surface protein expression of MRP5 on pancreatic cancer cells was assessed by staining with anti-MRP5 primary and control isotype IgG2a antibody. Fig. 1B and C show increased fluorescence staining with the MRP5 antibody (red colour) on MIA Paca-2 and PANC-1 cells (~three-fold) compared to the isotype control antibody (green colour), suggesting the expression of MRP5 protein on the surface of pancreatic cancer cells. Furthermore, the MRP5 efflux activity in MIA Paca-2 and PANC-1 cells was investigated by determining the cellular accumulation of a model MRP5 substrate, BCECF, in the presence or absence of MRP inhibitor benzbromarone at designated time-points. Benzbromarone significantly (P<0.0001) increased the steady-state accumulation of BCECF in both MIA Paca-2 and PANC-1 cells (Fig. 1D). Taken together, these results indicated that MIA Paca-2 and PANC-1 cells have high endogenous expression of MRP5.
Reduced MRP5 expression in MRP5-knockdown clones
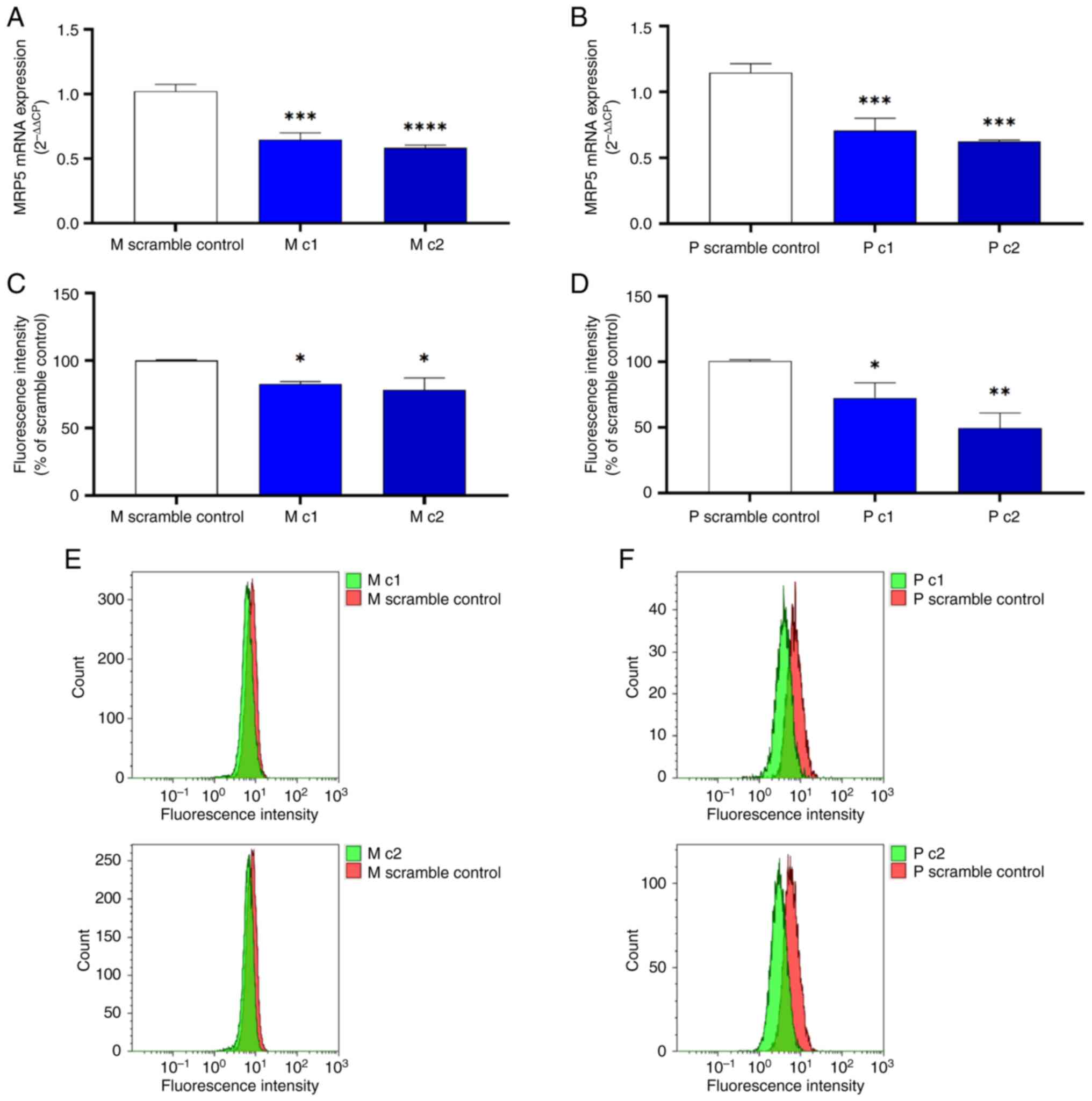
Significant reduction in the MRP5 mRNA expression was seen for all knockdown clones transduced with shRNA lentiviral particles. In MIA PaCa-2 cells, the mRNA transcripts of the MRP5 gene were significantly decreased by 37% (P<0.001) and 43% (P<0.0001) in M c1 and M c2 compared to the scramble control, respectively (Fig. 2A). A similar trend was seen in PANC-1 cells, as P c1 and P c2 showed a significant (P<0.001) reduction in MRP5 mRNA levels by 38 and 46% compared to the scrambled control, respectively (Fig. 2B).
MRP5 gene knockdown clones were further confirmed at the protein level by cell surface staining. The mean fluorescence intensity that indicates the MRP5 surface expression was decreased by 17 and 22% for M c1 (P<0.05) and M c2 (P<0.05) and by 28 and 51% for P c1 (P<0.05) and P c2 (P<0.01) compared to the scrambled control, respectively (Fig. 2C-F). These results suggested that transduction of shRNA lentiviral particles results in a significant decrease of the MRP5 mRNA and surface protein expression of both MIA PaCa-2 and PANC-1 cells.
The integration of plasmid and the potential off-target effects of MRP5-short-hairpin RNA were examined in MRP5 knockdown cells (Fig. S1). No significant differences of ABCB1 and ABCC2 mRNA expression were found in MRP5 knockdown cells compared to scramble control. However, the accumulation of BCECF remained at the same level between MRP5 knockdown cells and scramble control for both MIA Paca-2 and PANC-1 cells (Fig. S2). This may be caused by either single-cell variability or the loss of substrate specificity in the models used in the present study.
MRP5 knockdown sensitises pancreatic cancer cells to gemcitabine growth inhibition
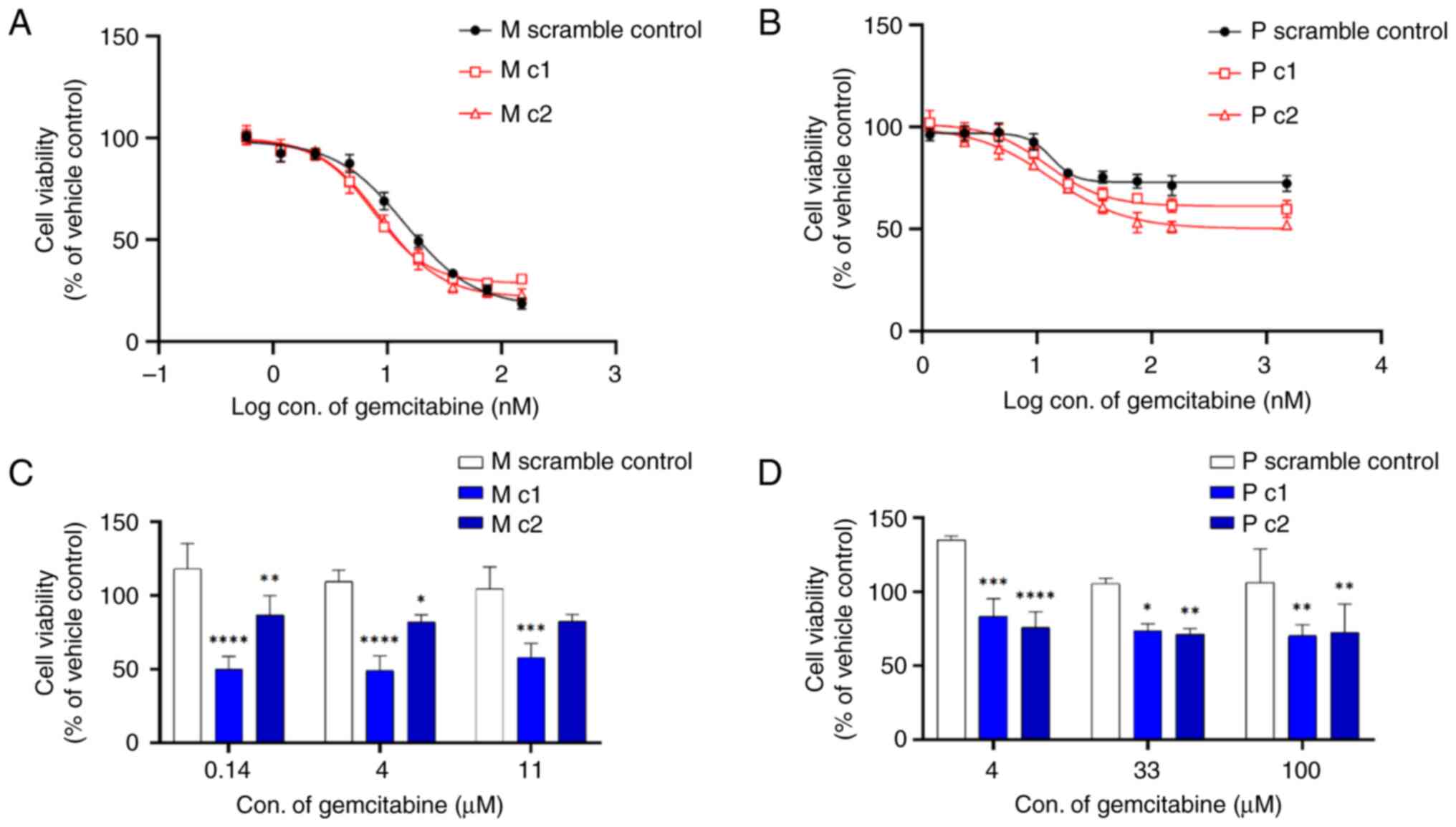
MIA PaCa-2 and PANC-1 clones were treated with 0–150 and 0–1,500 nM gemcitabine, respectively, in the 2D culture models. The 72-h cytotoxicity of gemcitabine was determined using an MTT assay. Gemcitabine inhibited the proliferation of scrambled control and knockdown clones in a dose-dependent manner. Table II shows that the IC50 values were decreased by 52% (M c1) and 42% (M c2) for MIA PaCa-2 cells (Fig. 3A) and by 46% (P c1) and 41% (P c2) for PANC-1 cells (Fig. 3B) compared to the scrambled control, respectively. Similar patterns were observed in pancreatic cancer cells by using colony formation assays (Fig. S3). The response of the two pancreatic cancer cell lines to gemcitabine was also examined in an in vitro 3D cell culture model. Gemcitabine was more potent against control pancreatic cancer cells (IC50 values of 17.26 and 13.88 nM for Mia PaCa-2 and PANC-1 cells, respectively) grown in 2D cultures in comparison with those in 3D cultures (both IC50 values >100 µM). Silencing MRP5 led to markedly increased sensitivity to gemcitabine in 3D culture models of both Mia PaCa-2 and PANC-1 compared to the control (Fig. 3C and D). In contrast, the knockdown clones and scramble control for both MIA Paca-2 and PANC-1 cells showed the same vulnerability to a poly (ADP-ribose) polymerase inhibitor olaparib, which is not an MRP5 substrate (Fig. S4 and Table SI).
Table II.Gemcitabine-induced growth inhibition: Comparison between scramble control and multidrug resistance protein 5-short-hairpin RNA-transduced clones in 2D cultures. |
MRP5 knockdown increases the gemcitabine-induced apoptosis rate
Gemcitabine-induced apoptosis was measured by detecting caspase activity in the apoptotic population. The time course was determined based on the cell type. MIA PaCa-2 clones exposed to gemcitabine exhibited extensive apoptosis after a 24-h incubation. PANC-1 clones showed minor apoptosis after 24 h of treatment; thus, the incubation time was increased to 48 h. In MIA PaCa-2 clones M c1 and M c2 treated with 100 nM gemcitabine, the apoptotic population increased by 67.50±16.40% (P<0.01) and 44.99±5.21% (P<0.01) compared to the scrambled control, respectively (Fig. 4A and B). After treatment with 200 nM gemcitabine, the apoptotic population increased by 53.15±32.50% (P<0.05) and 34.34±5.00% (P<0.05) for M c1 and M c2, respectively (Fig. 4A and B).
A similar trend was seen in PANC-1 cells-after treatment with 100 nM gemcitabine, the apoptotic population increased by 55.21±18.25 and 65.35±37.47% for P c1 (P<0.05) and P c2 (P<0.001), respectively (Fig. 4C and D). In PANC-1 cells treated with 200 nM gemcitabine, the apoptotic population increased by 97.18±23.44 and 49.09±20.94% for P c1 (P<0.05) and P c2 (P<0.05), respectively (Fig. 4C and D). These results signified increased gemcitabine sensitivity in ABCC5 knockdown pancreatic cancer cells.
MRP5 knockdown suppresses cell proliferation and migration
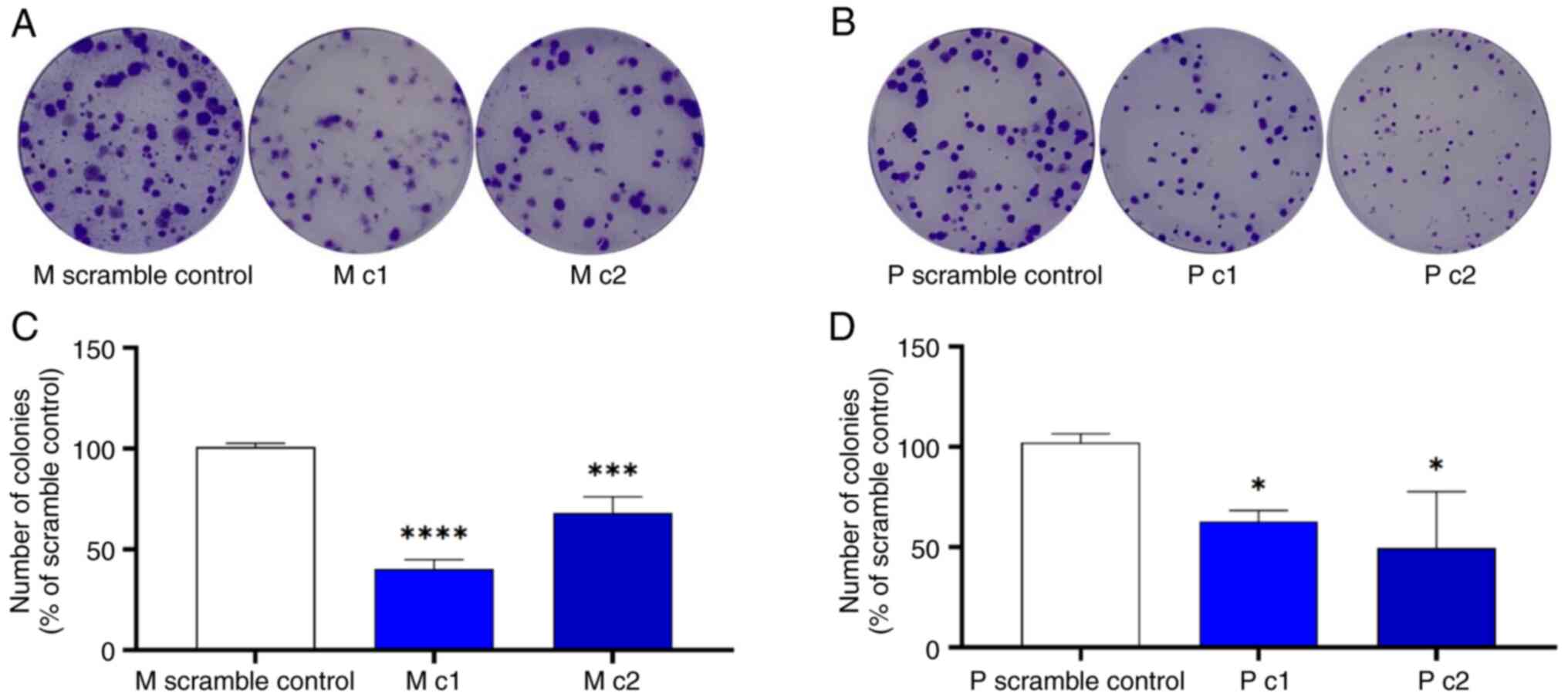
The effect of MRP5 on pancreatic cancer clonogenic capacity was examined using a colony-formation assay. Silencing of MRP5 decreased the clonogenic capacity of Mia PaCa-2 and PANC-1 cells (Fig. 5). The colonies formed within 10 days of incubation for the control Mia PaCa-2 and PANC-1 cells were significantly more numerous than those of the MRP5 knockdown cells.
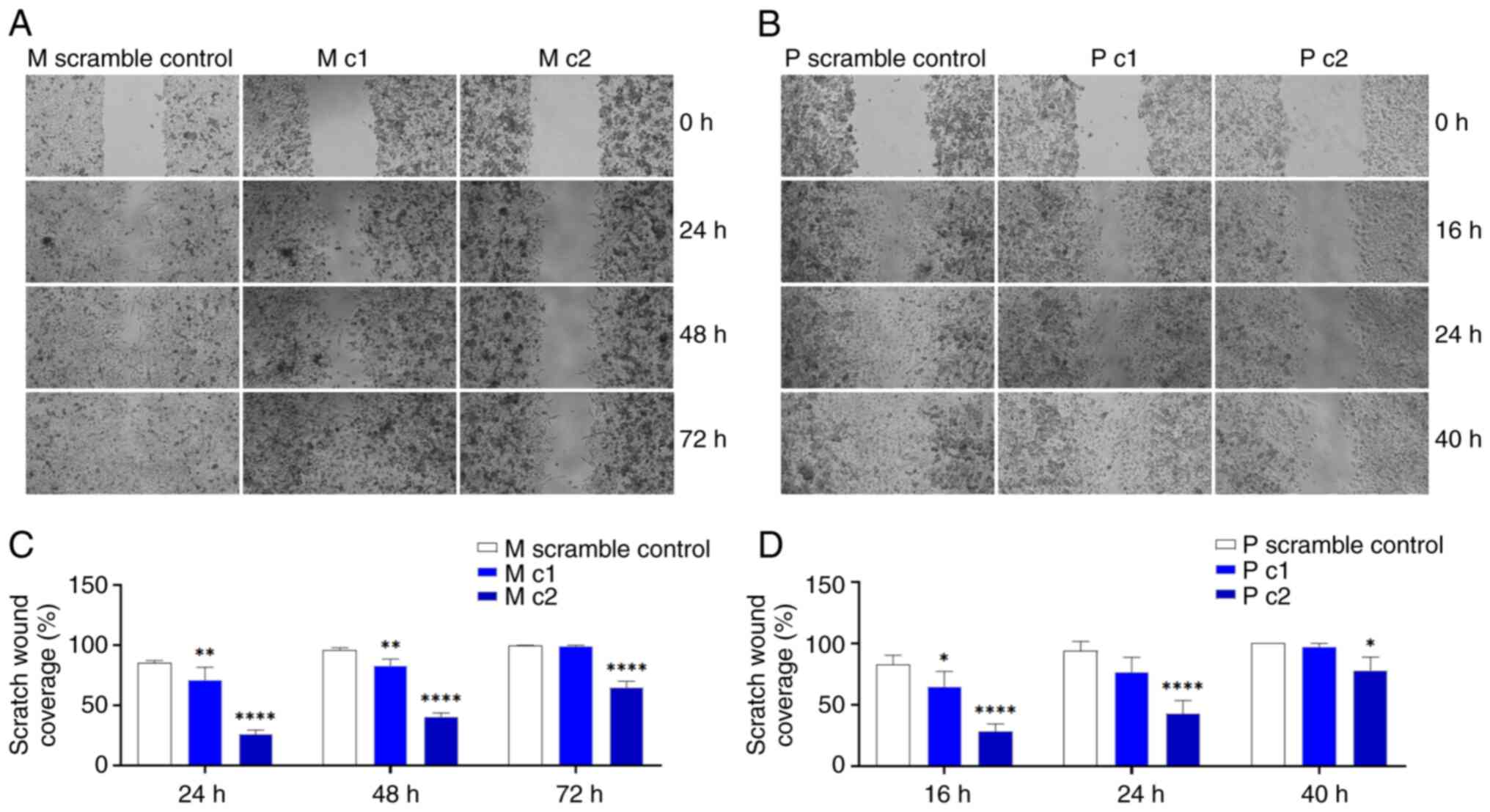
Silencing MRP5 decreased wound-induced migration in Mia PaCa-2 and PANC-1 cells (Fig. 6). Complete wound healing occurred within 72 and 40 h for the control Mia PaCa-2 and PANC-1 cells, respectively; but not in the MRP5 knockdown cell lines (Fig. 6A and B).
Silencing of MRP5 decreased cell migration in the Transwell assays quantifying cell migration toward the complete medium for 48 h (Fig. 7). The migrated cells per field were reduced by >50% for both cell lines examined.
Docking studies using AutoDock 4.2
In the present study, gemcitabine and cGMP were analysed through a molecular docking study using AutoDock 4.2 software. Docking results against receptor molecules obtained from Protein-Ligand Interaction Profiler showed that both gemcitabine and cGMP formed four hydrogen bonds with ABCC5. The estimated free energy of binding of gemcitabine and cGMP were determined to be −2.77 and −6.44 kcal/mol, respectively (Fig. 8).
Discussion
Based on robust evidence from multiple randomised controlled trials, gemcitabine and its combination regimens are clinically important for the treatment of pancreatic cancers. Although various patients benefit from gemcitabine-based treatment, certain patients fail to respond to treatment. There is an urgent need for developing strategies to overcome gemcitabine-based chemoresistance and improve the survival rate and life expectancy in the patients. Numerous factors are involved in the MDR of pancreatic cancer and one of the common factors is drug efflux via ABC transporters (44). Pancreatic cancer involves overexpression of numerous ABC transporter genes, such as MRP1, MRP2, MRP3, MRP4, MRP5, MRP8 and breast cancer resistance protein (BCRP) (4,45–47). These ABC transporters may work individually or together, which makes their mechanism of action quite intricate. Overexpression of these ABC transporters has also been correlated with resistance to chemotherapy. Accumulating evidence has shown that overexpression of MRP2, MRP3, MRP4 and MRP5 may be related to MDR and cancer progression (4,6,16,19,26,45–47).
Typical substrates of MRP5 include nucleotide analogues and gemcitabine is a nucleoside analogue. In vitro studies showed that MRP5 contributes to resistance to gemcitabine and other nucleoside analog drugs in PDAC cell lines and 293 cells overexpressing MRP5 (6,20,28,48). Exposure of PDAC cells to gemcitabine markedly increased MRP5 levels, thus indicating a drug-induced resistance mechanism of PDAC cells (28,48). Previous studies have revealed that the use of MRP5 inhibitor curcumin significantly enhanced cytotoxicity of gemcitabine on MRP5-overexpressing pancreatic cancer cells, which showed its reversal effect on MDR mediated by MRP5 (32,49). On the other hand, however, one study reported that overexpression of MRP4 and MRP5 conferred resistance to cytarabine and troxacitabine, but not gemcitabine (50). Such discrepancy may be due to the differences in MRP5 expression on the plasma membrane among different cell clones (11). Several studies examining the importance of other ABC transporters in gemcitabine resistance have confirmed that abnormal expression of P-glycoprotein, MRP1, MRP4 and BCRP is associated with MDR in pancreatic cancer (51,52). Some research has shown that MRP2 and MRP4 may have affinity for nucleotide analogues (6). Although in vitro evidence suggests that overexpression of ABC transporters confers resistance to cytotoxic and molecularly targeted chemotherapies, studies indicate that ABC transporters may also contribute to cancer development and metastasis independent of their efflux function (6,12,16,28,32,45). In the present study, silencing MRP5 by lentiviral shRNA was used to further clarify the contribution of MRP5 to pancreatic cancer proliferation, migration and gemcitabine cytotoxicity in Mia PaCa-2 and PANC-1 cells with endogenous MRP5 expression.
In the present study, MRP5 gene knockdown in MIA Paca-2 and PANC-1 cells was confirmed on both the mRNA and protein levels. The MRP5 mRNA transcripts and surface immunostaining were significantly lower in the shRNA lentiviral particles-transduced cells as compared to the scrambled control. MRP5 has been demonstrated to confer gemcitabine resistance in various in vitro cell culture models (6,20,28,48). While the current study focused on the non-efflux function of MRP5, the increased gemcitabine sensitivity may be used as additional evidence of MRP5 silencing, in conjugation with the surface staining and RT-qPCR results.
The cytotoxicity assay demonstrated that gemcitabine sensitivity for the knockdown clones was increased by roughly 2-fold for both MIA PaCa-2 and PANC-1 cells. By contrast, the knockdown clones did not show increased vulnerability to olaparib, which is not an MRP5 substrate. This supports the current hypothesis that overexpression of MRP5 is one of the major reasons for gemcitabine resistance in pancreatic cancer cells. The effect of the MRP5 knockdown on gemcitabine-induced apoptosis in pancreatic cancer cells was also tested. Silencing MRP5 by shRNA resulted in an increase in gemcitabine-induced caspase3/7-mediated apoptosis in MIA Paca-2 and PANC-1 cells. Further studies are required to better understand the molecular mechanisms of the potential roles of MRP5 in the protection against apoptosis in pancreatic cancer cells.
The present results suggest that depletion of MRP5 significantly reduced the number of colonies formed and diminished cell mobility/migration in pancreatic cancer cells. A recent study indicates that downregulation of MRP5 and BCRP induced cell apoptosis and reduced the migration ability of colorectal cancer cells (53). Upregulation of MRP5 was associated with enhanced cell proliferation, migration and invasion of prostate cancer in vitro and in vivo (54). It was previously reported that MRP5 is a mediator of breast cancer skeletal metastasis and loss of MRP5 expression diminishes the formation of breast cancer bone metastases without promoting breast cancer proliferation (55). It was postulated that the locally elevated level of cGMP, which is pumped out by MRP5, can further increase osteoclast motility. The in silico analysis of the present study indicated high binding affinity of cGMP towards MRP5, which supports the role of MRP5 in the accumulation of cGMP in the microenvironment. Whether pancreatic cancer cells that express MRP5, and thus maintain low cytoplasmic cGMP levels through active efflux, are associated with pancreatic cancer metastasis warrants further investigation.
Despite the coherence of the present results, there are several limitations to this study, including the low knockdown efficiency and potential off-target effects induced by shRNA-mediated gene silencing. More recently, the development of CRISPR inhibition and CRISPR activation has enabled researchers to explore the complex gene pathways associated with drug resistance and cancer hallmarks (56,57). Given the unique functionality of MRP5, such approaches would be applied to decipher the mechanism of actions of MRP5 and elucidate the regulatory pathways in pancreatic cancer. In addition, the present results were not validated in in vivo pancreatic cancer models, which offer complex biological systems and tumour microenvironment to evaluate cancer hallmarks and tumour resistance (58,59).
In conclusion, the present study adds to the growing body of evidence that MRP5 has an important role in cancer proliferation and migration independent of its drug efflux functions. Gemcitabine-based chemotherapy has been widely adopted as the standard and preferred chemotherapy regimen for treating advanced pancreatic cancer. Pancreatic cancer is highly metastatic and resistant to multiple drugs, including the front-line drug gemcitabine. The current results showed that MRP5 (ABCC5) confers gemcitabine resistance and promotes pancreatic cancer cell proliferation and migration, and modulation of MRP5 may reverse its protumour effect and enhance gemcitabine sensitivity. Therefore, it may be proposed that modulating MRP5 transporter activity or its regulatory pathways may be a potential therapeutic strategy in patients with high expression levels of MRP5.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was funded by the Faculty of Health and Environmental Science (Auckland University of Technology, NZ) Research and Development Fund 2022.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
Conceptualization, ZW and YL; methodology, JH, PB, JL, RB, SL, XY, FT and YL; software, JH, PB, JL, RB, SL and FT; validation, JH, PB, JL, RB and SL; formal analysis, JH and YL; investigation, JH, PB, JL, RB and SL; resources, YL; data curation, JH; writing - original draft preparation, JH and PB; writing - review and editing, JH, PB, JL, XY, FT, ZW and YL; visualization, JH and JL; supervision, YL; project administration, ZW and YL; funding acquisition, YL. JH and YL confirm the authenticity of all the raw data. All authors have read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Adamska A, Domenichini A and Falasca M: Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int J Mol Sci. 18:13382017. View Article : Google Scholar : PubMed/NCBI | |
|
Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et al: Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 369:1691–1703. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C, et al: FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 364:1817–1825. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Adamska A, Ferro R, Lattanzio R, Capone E, Domenichini A, Damiani V, Chiorino G, Akkaya BG, Linton KJ, De Laurenzi V, et al: ABCC3 is a novel target for the treatment of pancreatic cancer. Adv Biol Regul. 73:1006342019. View Article : Google Scholar : PubMed/NCBI | |
|
Gnanamony M and Gondi CS: Chemoresistance in pancreatic cancer: Emerging concepts. Oncol Lett. 13:2507–2513. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Adamska A and Falasca M: ATP-binding cassette transporters in progression and clinical outcome of pancreatic cancer: What is the way forward? World J Gastroenterol. 24:3222–3238. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Ozben T: Mechanisms and strategies to overcome multiple drug resistance in cancer. FEBS Lett. 580:2903–2909. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
O'Reilly EM: Pancreatic adenocarcinoma: New strategies for success. Gastrointest Cancer Res. 3:S11–S15. 2009.PubMed/NCBI | |
|
Vincent A, Herman J, Schulick R, Hruban RH and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C and Gottesman MM: Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 5:219–234. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Borst P, de Wolf C and van de Wetering K: Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch. 453:661–673. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE and Gottesman MM: Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 18:452–464. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Dvorak P, Pesta M and Soucek P: ABC gene expression profiles have clinical importance and possibly form a new hallmark of cancer. Tumour Biol. 39:10104283176998002017. View Article : Google Scholar : PubMed/NCBI | |
|
Sodani K, Patel A, Kathawala RJ and Chen ZS: Multidrug resistance associated proteins in multidrug resistance. Chin J Cancer. 31:58–72. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Tiwari AK, Sodani K, Dai CL, Ashby CR Jr and Chen ZS: Revisiting the ABCs of multidrug resistance in cancer chemotherapy. Curr Pharm Biotechnol. 12:570–594. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Bugde P, Biswas R, Merien F, Lu J, Liu DX, Chen M, Zhou S and Li Y: The therapeutic potential of targeting ABC transporters to combat multi-drug resistance. Expert Opin Ther Targets. 21:511–530. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Fukuda Y and Schuetz JD: ABC transporters and their role in nucleoside and nucleotide drug resistance. Biochem Pharmacol. 83:1073–1083. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Vasiliou V, Vasiliou K and Nebert DW: Human ATP-binding cassette (ABC) transporter family. Hum Genomics. 3:281–290. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Choi YH and Yu AM: ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 20:793–807. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Reid G, Wielinga P, Zelcer N, De Haas M, Van Deemter L, Wijnholds J, Balzarini J and Borst P: Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol. 63:1094–1103. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Konig J, Hartel M, Nies AT, Martignoni ME, Guo J, Büchler MW, Friess H and Keppler D: Expression and localization of human multidrug resistance protein (ABCC) family members in pancreatic carcinoma. Int J Cancer. 115:359–367. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Dazert P, Meissner K, Vogelgesang S, Heydrich B, Eckel L, Böhm M, Warzok R, Kerb R, Brinkmann U, Schaeffeler E, et al: Expression and localization of the multidrug resistance protein 5 (MRP5/ABCC5), a cellular export pump for cyclic nucleotides, in human heart. Am J Pathol. 163:1567–1577. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Mohelnikova-Duchonova B, Brynychova V, Oliverius M, Honsova E, Kala Z, Muckova K and Soucek P: Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas. 42:707–716. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L and Li J: Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 370:153–164. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Slot AJ, Molinski SV and Cole SP: Mammalian multidrug-resistance proteins (MRPs). Essays Biochem. 50:179–207. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng S, Pottler M, Lan B, Grützmann R, Pilarsky C and Yang H: Chemoresistance in pancreatic cancer. Int J Mol Sci. 20:45042019. View Article : Google Scholar : PubMed/NCBI | |
|
Tanaka M, Okazaki T, Suzuki H, Abbruzzese JL and Li D: Association of multi-drug resistance gene polymorphisms with pancreatic cancer outcome. Cancer. 117:744–751. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Hagmann W, Faissner R, Schnolzer M, Löhr M and Jesnowski R: Membrane drug transporters and chemoresistance in human pancreatic carcinoma. Cancers (Basel). 3:106–125. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Oguri T, Achiwa H, Sato S, Bessho Y, Takano Y, Miyazaki M, Muramatsu H, Maeda H, Niimi T and Ueda R: The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: A role of ABCC5 in gemcitabine sensitivity. Mol Cancer Ther. 5:1800–1806. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Fletcher JI, Haber M, Henderson MJ and Norris MD: ABC transporters in cancer: More than just drug efflux pumps. Nat Rev Cancer. 10:147–156. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Domenichini A, Adamska A and Falasca M: ABC transporters as cancer drivers: Potential functions in cancer development. Biochim Biophys Acta Gen Subj. 1863:52–60. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
He J, Fortunati E, Liu DX and Li Y: Pleiotropic roles of ABC transporters in breast cancer. Int J Mol Sci. 22:31992021. View Article : Google Scholar : PubMed/NCBI | |
|
Calcagno AM and Ambudkar SV: Analysis of expression of drug resistance-linked ABC transporters in cancer cells by quantitative RT-PCR. Methods Mol Biol. 637:121–132. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Li C, Chen D, Luo M, Ge M and Zhu J: Knockdown of ribosomal protein L39 by RNA interference inhibits the growth of human pancreatic cancer cells in vitro and in vivo. Biotechnol J. 9:652–663. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Liang CC, Park AY and Guan JL: In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al: Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al: AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50:D439–D444. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T and Hutchison GR: Open babel: An open chemical toolbox. J Cheminform. 3:332011. View Article : Google Scholar : PubMed/NCBI | |
|
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ and Schroeder M: PLIP 2021: Expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 49:W530–W534. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Schoning-Stierand K, Diedrich K, Fahrrolfes R, Flachsenberg F, Meyder A, Nittinger E, Steinegger R and Rarey M: ProteinsPlus: Interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 48:W48–W53. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Laskowski RA and Swindells MB: LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 51:2778–2786. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Kohan HG and Boroujerdi M: Time and concentration dependency of P-gp, MRP1 and MRP5 induction in response to gemcitabine uptake in Capan-2 pancreatic cancer cells. Xenobiotica. 45:642–652. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Amponsah PS, Fan P, Bauer N, Zhao Z, Gladkich J, Fellenberg J and Herr I: microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 388:107–117. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Kriebel PW, Majumdar R, Jenkins LM, Senoo H, Wang W, Ammu S, Chen S, Narayan K, Iijima M and Parent CA: Extracellular vesicles direct migration by synthesizing and releasing chemotactic signals. J Cell Biol. 217:2891–2910. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Biswas R, Bugde P, He J, Merien F, Lu J, Liu DX, Myint K, Liu J, McKeage M and Li Y: Transport-mediated oxaliplatin resistance associated with endogenous overexpression of MRP2 in Caco-2 and PANC-1 cells. Cancers (Basel). 11:13302019. View Article : Google Scholar : PubMed/NCBI | |
|
Nambaru PK, Hubner T, Kock K, Mews S, Grube M, Payen L, Guitton J, Sendler M, Jedlitschky G, Rimmbach C, et al: Drug efflux transporter multidrug resistance-associated protein 5 affects sensitivity of pancreatic cancer cell lines to the nucleoside anticancer drug 5-fluorouracil. Drug Metab Dispos. 39:132–139. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Yoshida K, Toden S, Ravindranathan P, Han H and Goel A: Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. 38:1036–1046. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Adema AD, Floor K, Smid K, Honeywell RJ, Scheffer GL, Jansen G and Peters GJ: Overexpression of MRP4 (ABCC4) and MRP5 (ABCC5) confer resistance to the nucleoside analogs cytarabine and troxacitabine, but not gemcitabine. Springerplus. 3:7322014. View Article : Google Scholar : PubMed/NCBI | |
|
Bergman AM, Pinedo HM, Talianidis I, Veerman G, Loves WJ, van der Wilt CL and Peters GJ: Increased sensitivity to gemcitabine of P-glycoprotein and multidrug resistance-associated protein-overexpressing human cancer cell lines. Br J Cancer. 88:1963–1970. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Polgar O and Bates SE: ABC transporters in the balance: Is there a role in multidrug resistance? Biochem Soc Trans. 33:241–245. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
El-Daly SM, Abo-Elfadl MT, Hussein J and Abo-Zeid MAM: Enhancement of the antitumor effect of 5-fluorouracil with modulation in drug transporters expression using PI3K inhibitors in colorectal cancer cells. Life Sci. 315:1213202023. View Article : Google Scholar : PubMed/NCBI | |
|
Ji G, He S, Huang C, Gong Y, Li X and Zhou L: Upregulation of ATP binding cassette subfamily c member 5 facilitates prostate cancer progression and enzalutamide resistance via the CDK1-mediated AR Ser81 phosphorylation pathway. Int J Biol Sci. 17:1613–1628. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Mourskaia AA, Amir E, Dong Z, Tiedemann K, Cory S, Omeroglu A, Bertos N, Ouellet V, Clemons M, Scheffer GL, et al: ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res. 14:R1492012. View Article : Google Scholar : PubMed/NCBI | |
|
Schmidt R, Steinhart Z, Layeghi M, Freimer JW, Bueno R, Nguyen VQ, Blaeschke F, Ye CJ and Marson A: CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science. 375:eabj40082022. View Article : Google Scholar : PubMed/NCBI | |
|
Morelli E, Gulla A, Amodio N, Taiana E, Neri A, Fulciniti M and Munshi NC: CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) to explore the Oncogenic lncRNA network. Methods Mol Biol. 2348:189–204. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Borst P: Looking back at multidrug resistance (MDR) research and ten mistakes to be avoided when writing about ABC transporters in MDR. FEBS Lett. 594:4001–4011. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Qi R, Bai Y, Li K, Liu N, Xu Y, Dal E, Wang Y, Lin R, Wang H, Liu Z, et al: Cancer-associated fibroblasts suppress ferroptosis and induce gemcitabine resistance in pancreatic cancer cells by secreting exosome-derived ACSL4-targeting miRNAs. Drug Resist Updat. 68:1009602023. View Article : Google Scholar : PubMed/NCBI |