Expression of PVRL4, a molecular target for cancer treatment, is transcriptionally regulated by FOS
- Authors:
- Published online on: December 5, 2023 https://doi.org/10.3892/or.2023.8676
- Article Number: 17
-
Copyright: © Nanamiya et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Breast cancer is the most common cancer affecting women and is one of the leading causes of cancer-related deaths in women (1). Although the mortality of breast cancer has decreased with the advancement of early detection and treatment (2), thousands of women die from this disease each year and the prognosis of patients with distant metastasis remains poor (2,3). Therefore, the development of new therapeutic strategies is a matter of pressing concern.
The PVRL4 gene encodes nectin-4, one of the nectin and nectin-like family calcium-independent cell adhesion molecules (4). This family consists of two subgroups, one containing nectin-1 to −4 that associate with afadin, a PDZ domain-containing cytoplasmic adaptor protein, and another containing nectin-like cell adhesion molecules, nectin-like-1 to −5 (5). Nectin-1 to −4 have an extracellular region containing a distal IgV-like domain, two IgC-like domains, a single transmembrane region and a cytoplasmic region with a C-terminal PDZ binding motif (6). Nectins are involved in cell adhesion by interacting with each other and/or other adhesion molecules, including cadherins, through their extracellular regions, and their cytoplasmic regions function as an anchor to the cellular cytoskeleton by binding with adaptor proteins such as afadin, PAR3 and band 4.1B (7). In addition, nectin-1 has been shown to act as a viral entry receptor for the human herpes simplex virus (8), and nectin-4 as a receptor for the measles virus (MV) (9). Although nectin-1, −2 and −3 are widely expressed in adult tissues, nectin-4 expression is restricted to fetal tissues and adult organs such as the throat and salivary gland (ducts), mammary gland and the skin (epidermis and sweat glands) (6). Notably, PVRL4 expression is elevated in a wide range of human cancer types such as breast, lung and ovarian cancer (10–13). Elevated PVRL4 expression confers the anchorage-independent proliferation of breast cancer cells, induction of integrin β4 signaling and subsequent Src family kinase activation in a matrix attachment-independent manner (14). In addition, nectin-4 overexpression in MDA-MB-231 cells, a nectin-4 null breast cancer cell line, induced epithelial-mesenchymal transition and metastasis, and upregulated the WNT/β-catenin signaling (15). Furthermore, PVRL4 could serve as a useful prognostic predictor of breast, lung, esophageal and high-grade T1 bladder cancer (12,16,17).
Notably, recent studies revealed that PVRL4 is a promising molecular target for the treatment of cancer (18–20). The antibody-drug conjugate, enfortumab vedotin, interacts with PVRL4 and is administered for the treatment of urothelial bladder cancer and other PVRL4+ solid tumors. The proliferation of human breast, bladder, pancreatic and lung cancer cells was significantly suppressed by treatment with enfortumab vedotin in mice xenograft models (21). Since PVRL4 is one of the known entry receptors for the MV, the application of oncolytic viruses may become another strategy for treatments that target PVRL4. Previously, Sugiyama et al (20) generated a recombinant MV HL-strain (rMV-SLAMblind) that carried a mutation in the region responsible for the interaction with signaling lymphocytic activation molecule (SLAM), another entry receptor for the MV. This genetically engineered virus efficiently infected breast cancer cells in a PVRL4-dependent fashion and decreased the viability of the cancer cells in vitro and in vivo, suggesting a therapeutic potential of rMV-SLAMblind as an oncolytic virus against human cancer expressing PVRL4.
Although expression of PVRL4 is elevated in a number of cancer types, including breast cancer (12,13,21), the mechanism of its induction in cancer cells remains to be elucidated. In addition, the global gene expression profile associated with PVRL4 has not yet, to the best of our knowledge, been clarified. Therefore, the aim of the present study was to identify the transcriptional regulator(s) of PVRL4 and to disclose the genes and pathways enhanced by PVRL4 overexpression in cancer cells. For this, regulatory regions within the PVRL4 gene were searched for using Assay for Transposase-Accessible Chromatin-sequencing (ATAC-seq) and chromatin immunoprecipitation-sequencing (ChIP-seq) data in combination with a reporter assay. In addition, candidate transcription factors whose binding motifs are localized in an enhancer region identified in the present study were further investigated. Furthermore, RNA-seq and subsequent pathway analyses were conducted using PVRL4-small interfering RNA (siRNA) in breast cancer cells expressing abundant PVRL4 to disclose the characteristics associated with its expression.
Materials and methods
Cell lines
Human breast cancer cell lines, SKBR3, T47D and MCF7, and human colorectal cancer cell lines, DLD1, LS174T, HCT116 and RKO, were obtained from the American Type Culture Collection. T47D and DLD1 cells were cultured in RPMI medium (Gibco; Thermo Fisher Scientific, Inc.), MCF7 and LST174T cells in EMEM (Gibco; Thermo Fisher Scientific, Inc.), SKBR3 and HCT116 cells in McCoy's 5A (Gibco; Thermo Fisher Scientific, Inc.) and RKO cells in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.), all containing 10% fetal bovine serum (Biosera, Ltd.) and antibiotic/antimycotic solution. The cells were incubated at 37°C under a humidified atmosphere containing 5% CO2. Mycoplasma contamination was tested using MycoStrip (Thermo Fisher Scientific, Inc.), which indicated that all cell lines were free of mycoplasma.
Reverse transcription-quantitative PCR (RT-qPCR)
Total cellular RNA was extracted from the cultured cells using a RNeasy Mini Kit (Qiagen, Inc.). cDNA was synthesized from 1 µg of total RNA with ReverTra Ace (Toyobo Life Science) according to the manufacturer's protocol. qPCR was performed using KAPA SYBR FAST qPCR Master Mix (2X) Kit (Sigma-Aldrich; Merck KGaA) with sets of primers for JUNB, JUN, FOS, JUND, activating transcription factor 3 (ATF3), ATF7, Jun dimerization protein 2 (JDP2), CAMP responsive element binding protein 1 (CREB1), CREB3L4, PVRL4 and hypoxanthine phosphoribosyltransferase 1 (HPRT1) (Table SI) and a StepOnePlus system (Thermo Fisher Scientific, Inc.). The thermocycler conditions were as follows: Denaturation at 95°C for 20 sec, followed by annealing and extension at 60°C for 20 sec, for a total of 40 cycles. Quantities of transcripts were determined by relative standard curves, and HPRT1 was used as the internal control. The quantification of the JUNB, JUN, FOS, JUND, ATF3, ATF7, JDP2, CREB1, CREB3L4, PVRL4 and HPRT1 transcripts were calculated from five-point standard curves prepared by amplifying the pooled control cDNA derived from SKBR3 cells according to Getting Started Guide of Applied Biosystems StepOne™ and StepOnePlus™ Real-Time PCR Systems Standard Curve Experiments (PN: 4376784; Thermo Fisher Scientific, Inc.). The standard curves were automatically generated by the StepOne Software v2.2.3. Baseline cycles and thresholds were set manually, and the quantification cycles were calculated automatically by the software using an in-built algorithm.
Gene silencing
Pools of human specific siRNAs were obtained from Merck KGaA (Table SII). ON-TARGETplus Non-targeting Pool (cat. no. D-001810-10; GE Healthcare Dharmacon, Inc.) was used as the control. All gene silencing experiments were performed in SKBR3 cells excepting si-FOS, where T47D cells were also used. SKBR3 and T47D cells were seeded 1 day before siRNA treatment. Cells were transfected with 10 nM siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.). After 48 h of incubation, total RNA was isolated from the cells using an RNeasy Mini Kit according to the manufacturer's instruction. The silencing effect was evaluated by quantitative RT-qPCR, as aforementioned.
Preparation of plasmids
Putative promoter regions in the 5′-flanking sequence, intron, and 3′-flanking sequence of PVRL4 were amplified by PCR with specific primer sets (Table SIII and IV) containing recognition sites for the following restriction enzymes: XhoI, BglII, KpnI or HindIII. PCR products were digested with restriction enzymes and cloned into the pGL4.23 vector (Promega Corporation). Mutant versions of PVRL4 reporter plasmids were generated by site-directed mutagenesis. Briefly, PCR was performed using KOD Plus NEO enzyme (Toyobo Life Science), 100 ng of the pGL4.23 -PVRL4#10-III plasmid containing the enhancer region as a template and a set of primers for Mut-1 and −2 (Table SV), according to the manufacturer's protocol. The PCR products were digested with Dpn1 restriction enzyme for 2 h at 37°C and were transformed into Escherichia coli DH5α cells.
To generate plasmids expressing FOS, the entire coding sequence of FOS was amplified by PCR using KOD ONE (Toyobo Life Science) with a set of primers (Table SVI) containing EcoRI or XhoI restriction sites, according to the manufacturer's protocol. The PCR product was cloned into a pCMV-myc vector (Promega Corporation). Sanger sequencing [using Applied Biosystems 3500 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol] confirmed the full-length cDNA sequence of FOS was inserted into the plasmid.
Luciferase assay
Cells seeded in 12-well plates were transfected with 0.3 µg of pGL4.23 reporter plasmid and 0.05 µg of pRL-TK plasmid (Promega Corporation) by FuGENE 6 reagent (Promega Corporation). After 48 h of transfection, a PicaGene Dual Sea Pansy Luminescence Kit (TOYOB-Net) was utilized to measure the activities of firefly and Renilla luciferase according to the supplier's protocol. The Renilla activity was normalized to the firefly activity.
Putative transcription factor binding site
To identify DNA sequences for putative transcription factor binding sites, ChIP-seq data from the ENCODE project [http://www.genome.ucsc.edu; The University of California Santa Cruz Genome Browser Database (UCSC); accession nos. GSM733646, GSM733674 and GSM733771] and JASPER (http://jaspar.genereg.net/) were used. In the present study, a JASPAR score >15.5 was deemed a putative-binding motif.
ATAC-Seq
ATAC-Seq of breast and colorectal cancer cells expressing either high or low levels of PVRL4 was performed using an ATAC-Seq Kit (Active Motif, Inc.), according to the manufacturer's instructions. Briefly, the cells were resuspended in 100 µl of ATAC lysis buffer to extract the nuclei. The nuclei were collected by centrifugation at 500 × g for 5 min at 4°C, resuspended in 50 µl of Tagmentation Master Mix (including assembled transposomes) and then incubated at 37°C for 30 min with shaking. The tagmented DNA was purified using 250 µl of DNA Purification Binding Buffer and 5 µl of 3 M sodium acetate. The DNA library was collected using SPRI beads, which binds to a magnet, and eluted with 20 µl of DNA Purification Elution Buffer. The quality of library was verified using a bioanalyzer (Agilent Technologies, Inc.). The libraries were sequenced (primers listed in Table SVII) with HiSeq Rapid SBS Kit v2 (Illumina, Inc) and HiSeq Rapid SR Cluster Kit v2 (Illumina, Inc) using 60 bp single-end reads on the HiSeq2500 platform (cat. no. SY-401-2501; Illumina, Inc). The raw sequencing reads were analyzed for quality using FastQC and then aligned to the human genome (hg19) using Bowtie2 (v2.4.1; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Integrative Genomics Viewer was used to visualize the sequencing data (https://igv.org/doc/desktop/).
Chromatin conformation capture (3C) assay
The 3C assay was performed as previously described (22,23). Briefly, SKBR3 cells were cross-linked with 1% formaldehyde for 10 min at room temperature and then quenched with 125 mM glycine. The cross-linked chromatin was digested at 37°C overnight with 400 U BgIII (Takara Bio, Inc.), which was then heat-inactivated for 25 min at 65°C with 1.6% SDS. Subsequently, DNA fragments were ligated with T4 DNA ligase (New England Biolabs, Inc.) for 8 h at 16°C. Samples were treated with 300 µg of proteinase K (Merck KGaA) at 37°C overnight to remove the cross-link and then with 300 µg of RNase A (Merck KGaA). The ligated DNA was purified by phenol/chloroform extraction and ethanol precipitation. The first PCR reaction was amplified with the outer primer sets. The first PCR parameters for relative quantification were as follows: 2 min at 98°C, followed by 35 cycles at 98°C for 10 sec, 60°C for 5 sec and 68°C for 10 sec. After the products of the first PCR were purified, nested PCR (KOD One; Toyobo Life Science) was performed using these products with the inner primer sets to investigate a possible interaction between the promoter and enhancer regions of PVRL4. The nested PCR parameters for relative quantification were as follows: 2 min at 98°C, followed by 30 cycles at 98°C for 10 sec, 62.5°C for 5 sec and 68°C for 10 sec. The sequences of the PCR primers used are shown in Table SVIII. The PCR products were confirmed by gel electrophoresis using 1.5% agarose gels and visualized with ethidium bromide staining (Sigma-Aldrich; Merck KGaA).
ChIP
The ChIP and ChIP-qPCR analysis were performed as previously described (24). A total of 2×107 SKBR3 cells were cross-linked with 1% formaldehyde for 10 min, followed by quenching with 125 mM glycine for 5 min. DNA fragmentation was performed using a UD-201 homogenizer (Tomy Seiko Co., Ltd.) with the following settings: Output level 5, 50% duty, 15 sec, 3 cycles and on floating ice to obtain 200–500 bp DNA fragments. The fragmented DNA samples were confirmed by gel electrophoresis using 1.5% agarose gels and visualized with ethidium bromide staining (Sigma-Aldrich; Merck KGaA). An aliquot of the sample was kept as an input and the remaining sample was used for ChIP analysis. The fragmented samples from the SKBR cells were incubated with 8 µg of anti-phospho-FOS antibody (Cell Signaling Technology, Inc.; cat. no. 5348; 1:100) or 8 µg of anti-IgG antibody (normal mouse IgG; Santa Cruz Biotechnology, Inc.; cat. no. sc-2025; 1:400) and bound to protein G-sepharose beads (GE Healthcare) at 4°C overnight. The beads were separated with a column and washed sequentially for 5 min with 1 ml of the following buffers: 1X low salt wash buffer [1% Triton X-100, 1 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl (pH 8.0)], 1X high salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl (pH 8.0)], 1X LiCl wash buffer [0.25 M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl (pH 8.0)] and 2X Tris EDTA buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA]. The beads were then eluted with 200 µl of elution buffer (100 mM NaHCO3, 1% SDS, 5 mM NaCl) and 2 µl of proteinase K (10 mg/ml; Sigma-Aldrich; Merck KGaA). The immunoprecipitated and input DNA were purified by using a QIAquick PCR Purification Kit (Qiagen, Inc.). The concentration of two purified DNA samples was measured using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Inc.) with a Qubit fluorometer (Thermo Fisher Scientific, Inc.). qPCR was performed using KAPA SYBR FAST qPCR Master Mix (2X) Kit (Sigma-Aldrich; Merck KGaA) with primers shown in Table SIX. Amplification of the glyceraldehyde-3-phosphate dehydrogenase gene was used as a negative control, and relative quantification was performed using StepOne Software v2.2.3. The PCR parameters for relative quantification were as follows: 2 min at 98°C, followed by 40 cycles at 95°C for 1 sec and 60°C for 20 sec.
RNA-seq and Gene Ontology (GO) analysis
To identify genes regulated by PVRL4, RNA-seq analysis was performed using an Ion Proton system™ (Thermo Fisher Scientific, Inc.). Total RNA was extracted from SKBR3 cells treated with siPVRL4#1, #2 or control siRNA using a RNeasy Mini Kit (Qiagen, Inc.), and the quality of RNA was assessed using an Agilent bioanalyzer device (Agilent Technologies, Inc). Libraries were prepared using all samples that had a RNA integrity number >7.0. RNA-seq libraries were prepared with 100 ng total RNA using the Ion AmpliSeq Transcriptome Human GeneExpression Kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The libraries were sequenced using an Ion Proton system with an Ion PI Hi-Q Sequencing 200 kit and Ion PI Chip v3 (Thermo Fisher Scientific, Inc.). The sequencing reads were aligned to hg19_AmpliSeq_Transcriptome_ERCC_v1 using the Torrent Mapping Alignment Program (https://github.com/iontorrent/TMAP), and the raw count data were generated using the AmpliSeqRNA plug-in (v5.2.0.3), both from the Torrent Suite Software (v5.2.2; Thermo Fisher Scientific, Inc.). The DESeq2 package (v1.26.0; http://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to normalize the read count data and test for differential gene expression. False discovery rate-adjusted P-values (q-values) <0.5 were considered to indicate a statistically significant difference. Functional enrichment analysis was performed using Metascape (https://metascape.org/gp/index.html#/main/step1) (25). Metascape pathway enrichment analysis uses GO, Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/kegg_ja.html), Reactome (https://reactome.org/) and MSigDB (https://www.gsea-msigdb.org/gsea/msigdb). In brief, pairwise similarities between two enriched terms were calculated based on a κ test score. Then the similarity matrix was clustered hierarchically, and a similarity threshold score of 0.3-κ test score was applied to trim the result into separate clusters.
Statistical analysis
Unpaired two-tailed Student's t-test and one-way ANOVA followed by Dunnett's test were applied for statistical analyses. P<0.05 was considered to indicate a statistically significant difference. Data obtained from three independent experiments are presented as the mean ± SD.
Results
Candidate regulatory regions of PVRL4
It was reported that PVRL4 expression was elevated in ~61% of ductal carcinomas of the breast (11). However, genetic amplification of the PVRL4 gene was observed in ~10% of breast invasive carcinoma according to The Cancer Genome Atlas Pan Cancer Atlas data (https://gdc.cancer.gov/about-data/publications/pancanatlas), suggesting that different mechanisms may play a role in the elevated PVRL4 expression in breast cancer tissues.
To clarify the regulatory mechanisms of its expression in cancer cells, transcriptional regulatory elements in the PVRL4 genomic region were searched for. Analysis of the histone modifications, H3K4me1 and H3K27ac, in the UCSC genome database identified seven regions high in H3K4me1 and five regions high in H3K27ac, and the five high H3K27ac-regions overlapped with the high H3K4me1-regions (Figs. 1A and S1A). A further search of the database identified five DNase high sensitivity regions within the overlapped high H3K4me1 and high H3K27ac regions. To identify open chromatin regions, ATAC-seq was performed using breast cancer cells expressing high levels of PVRL4 (SKBR3 and T47D) and colon cancer cells expressing low levels of PVRL4 (HCT116 and RKO) (Fig. S1B). Subsequently, four putative open chromatin regions were identified with high ATAC-seq peaks in SKBR3 and T47D cells but not in HCT116 and RKO cells (Fig. 1B). Of these four regions, two were located in intron 4 and intron 6 and the remaining two were in the 3′-flanking region. Notably, three of the four regions were localized in the overlapped high H3K4me1 and high H3K27ac regions.
Identification of a transcriptional regulatory region in PVRL4
To identify the enhancer region within PVRL4, 14 regions were selected from the high H3K4me1 regions (Fig. 1A) and cloned into the pGL4.23 reporter plasmid. Transcriptional regulation is restricted by the physical constraints imposed by topologically associating domain (TAD). It is of note that these 14 regions were localized within the same TAD of the PVRL4- transcription start site according to the Topologically Associating Domain Knowledge Base (http://dna.cs.miami.edu/TADKB/), suggesting that the candidate regions may physically associate with the PVRL4 promoter region (Fig. S2). To examine the transcriptional activity of the candidate regions, a dual-luciferase assay using the 14 reporter plasmids was conducted in SKBR3 and T47D cells. The reporter plasmids containing regions #4, #9, #10 or #13 and those containing region #10 or #13 showed significantly higher luciferase activities compared with the control (mock) plasmids in SKBR3 and T47D cells, respectively (P<0.01; Figs. 1C and S3A). Since region #13 was localized in intron 1 of the Rho GTPase activating protein 30 (ARHGAP30) gene, this region was excluded from further analysis. Region #10 became the focus of further study as it demonstrated the highest transcriptional activity among the candidate regions. To elucidate the important region within region #10, three deletion mutants of #10 reporter plasmids (#10-I, #10-II and #10-III) were prepared and their reporter activities were compared with that of wild-type #10 reporter plasmids (Figs. 1D and S3B). Notably, deletion of the 5′-region (#10-I) increased the reporter activity in both SKBR3 and T47D cells. Furthermore, reporter plasmids containing #10-III also had a higher activity than the wild-type #10 plasmid in both cell lines, suggesting that this region may include transcriptionally important elements. Since the #10-III region was within an overlapping region (with high H3K4me1, high H3K27ac, high DNase sensitivity and open chromatin), it may play a role in the transcription of PVRL4 as a distant enhancer region.
Involvement of two putative FOS-binding motifs in the enhancer region
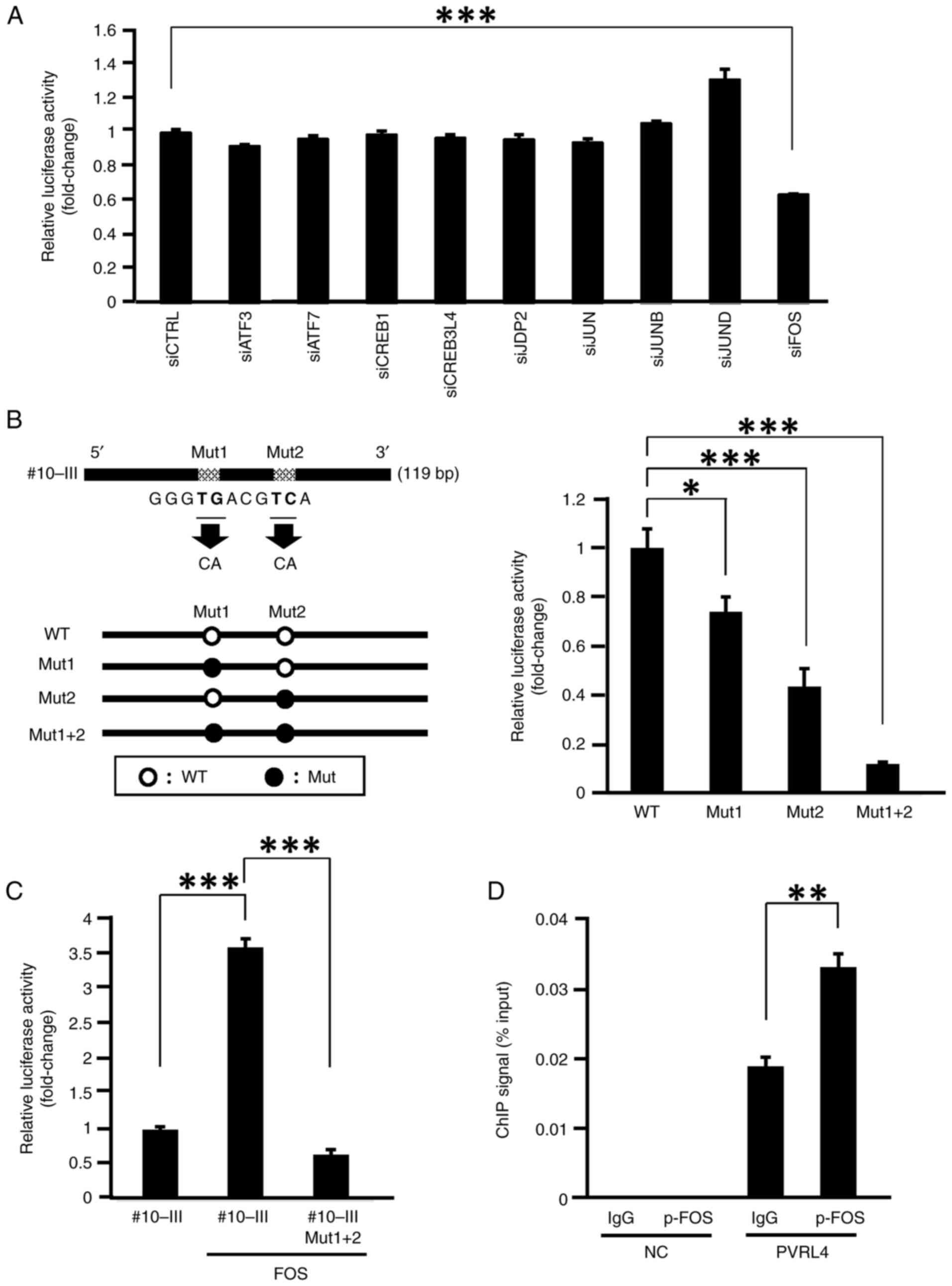
Using the ChIP-seq data from the ENCODE project, transcription factors that may interact with the candidate enhancer region (#10-III) and putative-binding motifs were searched for using the JASPER database. Subsequently, 11 candidate binding-motifs of transcription factors were identified (Table I). Since FOSB and FOS-like 2 (FOSL2) act as heterodimers with JUN, knockdown of JUN by siRNA will decrease the activity of the FOSB-JUN and FOSL2-JUN complexes (26). However, FOS is known to function both as a homodimer and a heterodimer with JUN (27). Therefore, FOSB and FOSL2 were excluded from the subsequent knockdown experiments and the effect of the other nine transcription factors (JUN, JUNB, JUND, JDP2, FOS ATF3, ATF7, CREB1 and CREB3L4) were examined. siRNAs targeting the nine transcription factors were generated and the knockdown effect of each siRNA on the expression of its target gene was confirmed by qPCR analysis (Fig. S4). The effect of each siRNA on the reporter activity of plasmids containing #10-III was then investigated in SKBR3 cells. FOS siRNA significantly reduced the reporter activity (P<0.005), but the remaining eight siRNAs did not have a significant effect (Fig. 2A). Since the #10-III region contained a putative FOS binding motif (TGACGTCA), mutant reporter plasmids carrying nucleotide substitutions in the binding motif (mut1, CAACGTCA; mut2, TGACGCAA; mut1+2, CAACGCAA) were constructed (Fig. 2B). As expected, these substitutions significantly reduced the reporter activity compared with the wild-type plasmid (pGL4.23-PVRL4#10-III) in SKBR3 cells (Figs. 2B and S5A).
To confirm that FOS transcriptionally regulates the expression of PVRL4, the reporter activity of pGL4.23-PVRL4#10-III was analyzed in the presence and absence of a plasmid expressing wild-type FOS (pCMV-FOS; efficiency of the overexpression of FOS is shown in Fig. S5B) in SKBR3 cells. As shown in Fig. 2C, the reporter activity increased 3.58-fold in the presence of pCMV-FOS. This increased reporter activity by pCMV-FOS was not observed with a mutant reporter plasmid containing two substitutions in the FOS-binding motifs (Mut1 + 2; Fig. 2C and S5C). Furthermore, the ChIP-qPCR assay demonstrated that the immunoprecipitation with p-FOS antibody enriched the DNA of region #10-III compared with the control IgG antibody (Fig. 2D), which is in agreement with the ENCODE results. These results suggested that FOS transcriptionally upregulated PVRL4 through its interaction with two FOS-binding motifs in intron 4.
Interaction of the enhancer region with the promoter of PVRL4
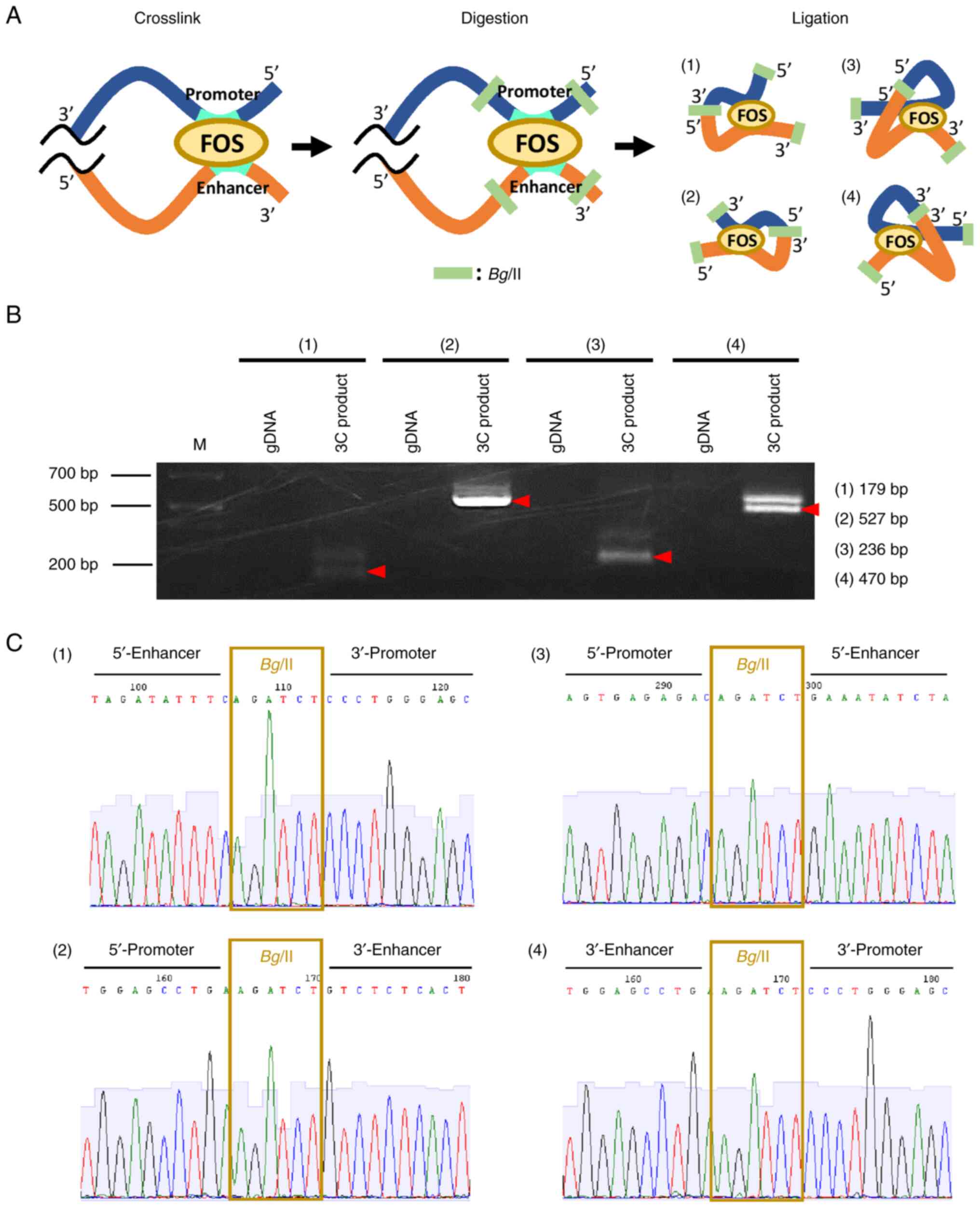
To examine whether the enhancer region interacts with the PVRL4 promoter, a 3C assay was conducted. DNA from SKBR3 cells was cross-linked with formaldehyde and subsequently digested with a restriction enzyme, Bglll. Self-ligation of the DNA was expected to produce chromatin loops between the enhancer and promoter regions when the two were closely associated (Fig. 3A). In total, four sets of first and nested PCR primers were designed that could detect associations between the two regions (Table SVII). Subsequently, amplification of the 3C DNA with the four primer sets produced PCR products with the expected sizes, but the amplification of control SKBR3 DNA failed to produce PCR products (Fig. 3B). Additionally, sequence analysis of the nested PCR products was conducted with primers in Table SVIII, which confirmed a ligated DNA sequence of the enhancer region (#10-III) and the promoter region (Fig. 3C). These data suggested that the enhancer interacted with the promoter region through the formation of a chromatin loop.
FOS is involved in PVRL4 expression
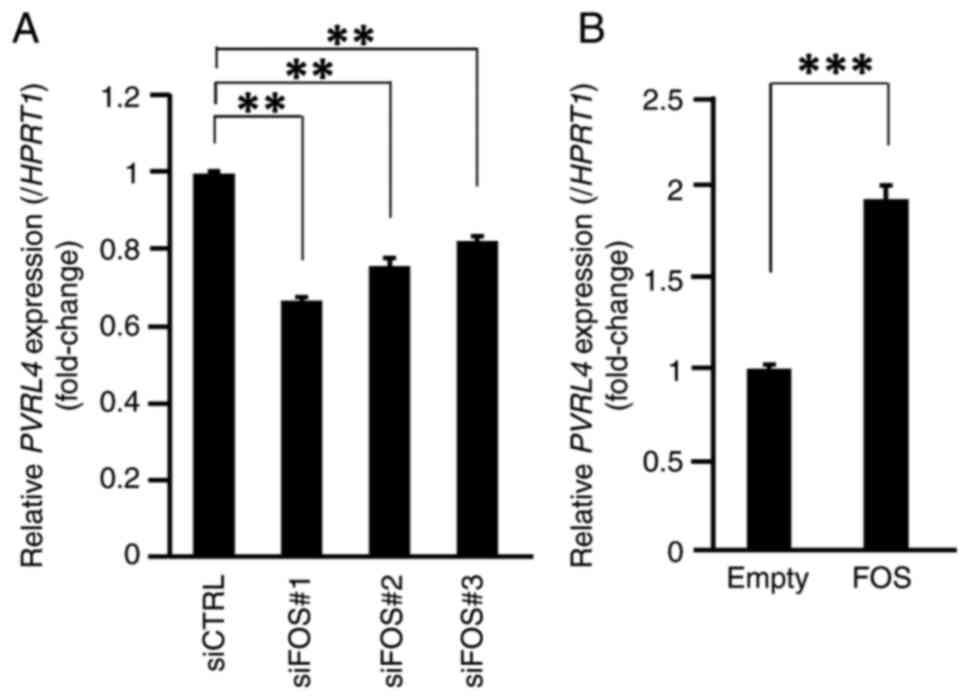
To investigate the involvement of FOS in the regulation of PVRL4, the effect of FOS knockdown on PVRL4 expression in SKBR3 and T47D cells was analyzed by qPCR. The efficiency of FOS knockdown by FOS siRNA transfection is shown in Fig. S4I and J. The expression of PVRL4 was significantly decreased by transfection with the three different FOS siRNAs (siFOS#1, #2 and #3; P<0.01; Figs. 4A and S6).
To confirm the effect of FOS on PVRL4 expression, MCF7 cells were transfected with pCMV-FOS and the expression of PVRL4 was analyzed by qPCR. Consistent with the reporter assay, PVRL4 expression was significantly enhanced by the overexpression of FOS (P<0.005; Fig. 4B).
Expression analysis suggested a link between PVRL4 with the immune system and apoptosis
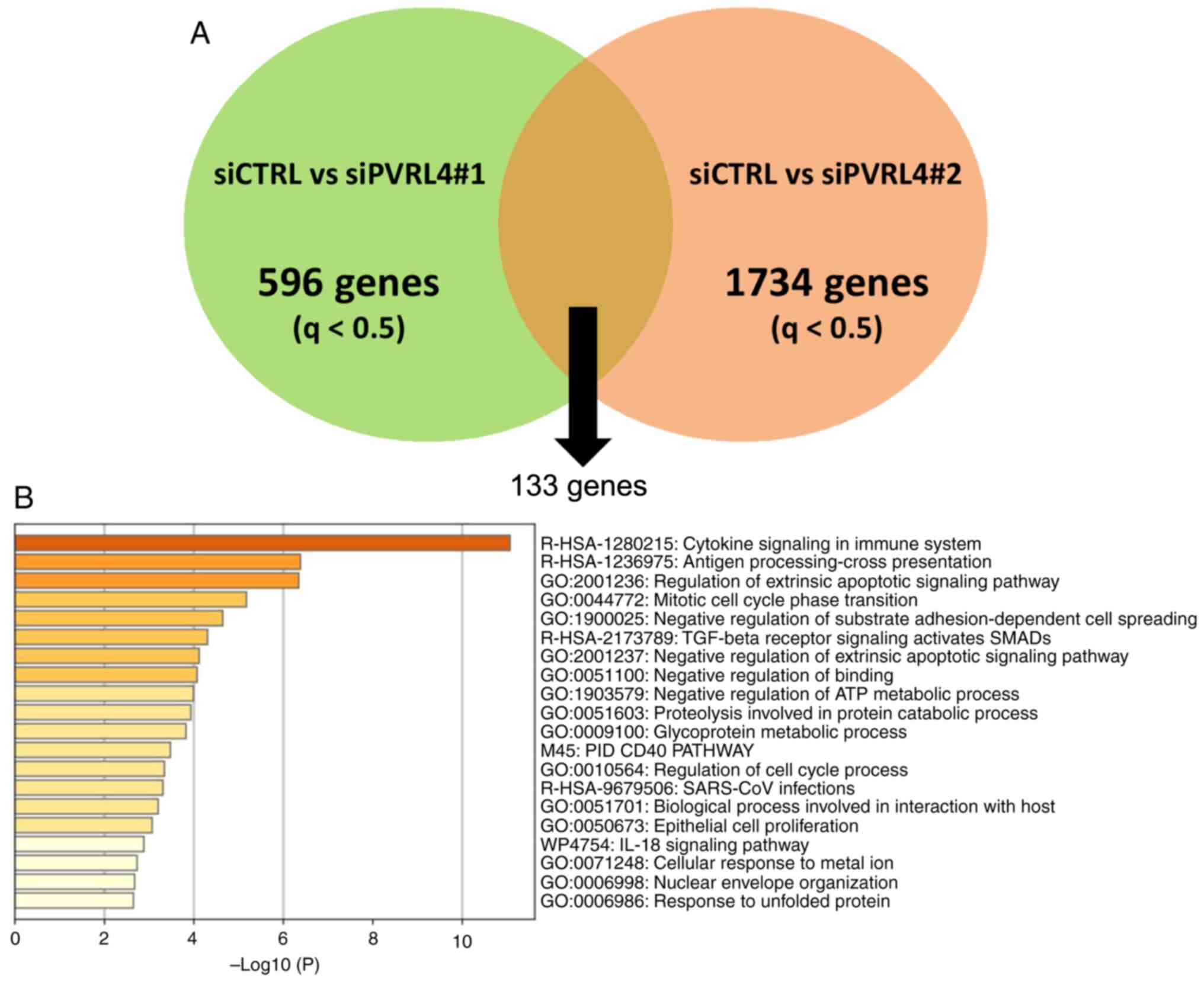
To clarify the function of PVRL4 in breast cancer cells, RNA-seq analysis using SKBR3 cells treated with control or PVRL4 siRNA (siPVRL4#1 and siPVRL4#2) was performed. The efficiency of PVRL4 knockdown following transfection with these siRNA is shown in Fig. S7A. A total of 596 and 1734 genes were identified whose expression levels were significantly altered by siPVRL4#1 and siPVRL4#2, respectively, compared with control siRNA (q<0.5; Figs. 5A and S7B). The number of genes was low since PVRL4 is a cell membrane receptor (4). The effect of these two siRNA on expression may be small compared with the siRNA of transcription factors. Since the inclusion of unmerged genes may have increased the likelihood of detecting GO of off-target effects, 133 genes that were commonly altered by the two siRNAs were used. GO analysis with the 133 genes found three ontology terms, ‘Cytokine Signaling in Immune System’, ‘Antigen processing-Cross presentation’ and ‘Regulation of Extrinsic Apoptotic Signaling Pathway’ (Fig. 5B). In both siPVRL4 treatments, the expression levels of IFIT1, IFI44, IFI44L, MX1, XAF1 and OAS2, related to ‘Cytokine Signaling in Immune System’, were upregulated 11.4–2.9-fold, compared with the control siRNA (Table SX). These six, IFIT1 (28), IFI44 (29), IFI44L (29), MX1 (30,31), XAF1 (30,32) and OAS2 (30,33), genes are known for their involvement in the defense against virus infection. Since PVRL4 is a member of the nectin family, altered expression of the genes involved in ‘Negative Regulation of Binding’ may suggest a decrease in cell adhesion. These results suggested that PVRL4 was involved in the cytokine response and immune system.
Discussion
In the present study, a distant enhancer region of PVRL4 in intron 4 was identified, and it was clarified that FOS was involved in the transcriptional regulation of PVRL4 through interaction with this enhancer region. Additionally, it was demonstrated that PVRL4 may downregulate the expression of genes associated with cytokine signaling and the immune system. In general, gene expression is regulated by several regions and a number of factors. Therefore, expression of PVRL4 may be regulated not only by the enhancer region but also by other regions. Moreover, the expression of PVRL4 may be controlled partially by FOS and other undetermined transcription factors.
A previous study revealed that c-FOS protooncogene expression was induced by estrogen in MCF7 breast cancer cells (34). Recently, Binato et al (35) reported that c-FOS and c-JUN proteins are induced in luminal A-type breast cancer cells, and that nuclear receptor-interacting protein 1 (NRIP1) was consequently augmented by the complex. Furthermore, it was found that expression levels of the progesterone receptor, estrogen receptor 1 and cyclin D1 were upregulated by NRIP1, suggesting a link between c-FOS and the proliferation of breast cancer cells (35). In addition, c-FOS is transcriptionally induced by ETS Transcription Factor ELK1 in bladder cancer (36). However, it remains to be clarified how frequently transcriptional activity of c-FOS is enhanced in different types of cancer cells. To transactivate downstream genes, FOS forms a dimeric complex with various dimer partners, such as JUN family proteins (c-JUN, JUNB and JUND), and the complex binds to the so-called TPA-responsive element (TGAC/GTCA) in the downstream genes through its leucine zipper structure (37). In addition to this heterodimerization, the activity of FOS is modulated through its phosphorylation by kinases, including ERK1/2 (38) and RSK1/2 (39). Although it was shown in the present study that FOS plays a crucial role in the expression of PVRL4, the involvement of dimer partner proteins in the induction of expression remains to be clarified. Since the regulatory mechanism of FOS-mediated transcriptional activity is complicated, further investigation is necessary.
A recent study reported that estrogen-related receptor-α (ESRRA) transcriptionally upregulates PVRL4 expression through an interaction with estrogen responsive elements in its promoter region (40). Although enhanced FOS expression is not frequently observed in breast cancer cells (26), the activity of FOS-heterodimers may be enhanced by its partner proteins or by post-transcriptional modifications of FOS protein. In addition, PVRL4 may be transcriptionally regulated by ESRRA and c-FOS in breast cancer cells, but this requires further experimental validation.
In the present study, RNA-seq and subsequent pathway enrichment analyses revealed that PVRL4 expression was associated with cytokine responses, antigen processing-cross presentation and the immune system. These results were in agreement with the report that PVRL4 functions as a ligand of TIGIT, the inhibitory receptor T-cell immunoreceptor with Ig and ITIM domains, and that PVRL4 inhibits the activity of natural killer cells (41). In addition, the cytoplasmic region of PVRL4 is involved in the interaction with the actin cytoskeleton through afadin. It was also reported that PVRL4 activates the JAK-STAT signaling pathway through association with suppressor of cytokine signaling 1 (SOCS1) (42,43). Therefore, in addition to TIGIT-mediated escape from the immune checkpoint, PVRL4 expression may mitigate cytokine signaling through the recruitment of SOCS1 and facilitate cells in suppressing immune responses. If these hypotheses are correct, decreased expression of PVRL4 and/or inhibition of PVRL4-mediated immune suppression may enhance the efficacy of immune checkpoint inhibitors. It is of note that IFIT1, IFI44, IFI44L, MX1, XAF1 and OAS2, the six genes upregulated by the knockdown of PVRL4, are expected to be downregulated in cells expressing PVRL4. Since these proteins are known to exhibit antiviral activity through inhibition of viral replication and the stabilization of antiviral immunity, PVRL4 (the MV receptor) expression may serve not only in the entry of the MV but also provide a suitable environment for their replication by suppressing antiviral reactions. Therefore, the development of new therapeutic modalities to suppress the expression of PVRL4 may contribute to efficient treatment for neoplasms expressing abundant PVRL4 as well as the symptoms caused by the infection of MV.
The limitations of the present study include the absence of tissue-specific control of PVRL4. Since breast cancer cell lines were used in the present study, PVRL4 regulatory mechanisms in other tissues may have been missed. As such, future studies may elucidate tissue-specific regulatory mechanisms of PVRL4 expression. In addition, the identified enhancer region, #10-III, may affect the expression of ARHGAP30 or other genes (44), which should be independently determined in future studies.
In conclusion, in the present study, it was determined that FOS directly regulated the transcriptional activity of PVRL4 in breast cancer cell lines. These results may assist with understanding the regulatory mechanism of PVRL4 and may contribute to the development of new strategies for cancer treatment and measles infection.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
We are grateful to research assistants: Ms. Seira Hatakeyama, Ms. Rika Koubo, and Ms. Yumiko Isobe (Division of Clinical Genome Research, The Institute of Medical Science, The University of Tokyo) for their technical assistance.
Funding
This study was supported in part by Health and Labour Sciences Research Grants of Japan (grant no. 15ck0106001h0003) and Japan Agency for Medical Research and Development (grant no. 19ck0106281h0003).
Availability of data and materials
The datasets generated or analyzed during the current study are available in the NCBI Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE236275 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE240039.
Authors' contributions
TN and YF designed the studies, and TN, KT, KY, MA and AS performed the experiments. TN and KT confirm the authenticity of all the raw data. TN, KY and TI provided analysis and interpretation of data. TN, KT, TF and YF wrote the manuscript. YO, TF, MY and CK contributed to data collection and interpretation, and critically reviewed the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Siegel RL, Miller KD and Jemal A: Cancer statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Narod SA, Iqbal J, Giannakeas V, Sopik V and Sun P: Breast cancer mortality after a diagnosis of ductal carcinoma in situ. JAMA Oncol. 1:888–896. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Chung CT and Carlson RW: Goals and objectives in the management of metastatic breast cancer. Oncologist. 8:514–520. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Duraivelan K and Samanta D: Emerging roles of the nectin family of cell adhesion molecules in tumour-associated pathways. Biochim Biophys Acta Rev Cancer. 1876:1885892021. View Article : Google Scholar : PubMed/NCBI | |
|
Samanta D and Almo SC: Nectin family of cell-adhesion molecules: Structural and molecular aspects of function and specificity. Cell Mol Life Sci. 72:645–658. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Chatterjee S, Sinha S and Kundu CN: Nectin cell adhesion molecule-4 (NECTIN-4): A potential target for cancer therapy. Eur J Pharmacol. 15:1745162021. View Article : Google Scholar : PubMed/NCBI | |
|
Ooshio T, Fujita N, Yamada A, Sato T, Kitagawa Y, Okamoto R, Nakata S, Miki A, Irie K and Takai Y: Cooperative roles of Par-3 and afadin in the formation of adherens and tight junctions. J Cell Sci. 120:2352–2365. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ishino R, Kawase Y, Kitawaki T, Sugimoto N, Oku M, Uchida S, Imataki O, Matsuoka A, Taoka T, Kawakami K, et al: Oncolytic virus therapy with HSV-1 for hematological malignancies. Mol Ther. 29:762–774. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, et al: Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 480:530–533. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Athanassiadou AM, Patsouris E, Tsipis A, Gonidi M and Athanassiadou P: The significance of survivin and nectin-4 expression in the prognosis of breast carcinoma. Folia Histochem Cytobiol. 49:26–33. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Fabre-Lafay S, Monville F, Garrido-Urbani S, Berruyer-Pouyet C, Ginestier C, Reymond N, Finetti P, Sauvan R, Adélaïde J, Geneix J, et al: Nectin-4 is a new histological and serological tumour associated marker for breast cancer. BMC Cancer. 7:732007. View Article : Google Scholar : PubMed/NCBI | |
|
Takano A, Ishikawa N, Nishino R, Masuda K, Yasui W, Inai K, Nishimura H, Ito H, Nakayama H, Miyagi Y, et al: Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res. 69:6694–6703. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Derycke MS, Pambuccian SE, Gilks CB, Kalloger SE, Ghidouche A, Lopez M, Bliss RL, Geller MA, Argenta PA, Harrington KM and Skubitz AP: Nectin 4 overexpression in ovarian cancer tissues and serum: Potential role as a serum biomarker. Am J Clin Pathol. 134:835–845. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Pavlova NN, Pallasch C, Elia AE, Braun CJ, Westbrook TF, Hemann M and Elledge SJ: A role for PVRL4-driven cell-cell interactions in tumorigenesis. Elife. 30:e003582013. View Article : Google Scholar : PubMed/NCBI | |
|
Siddharth S, Goutam K, Das S, Nayak A, Nayak D, Sethy C, Wyatt MD and Kundu CN: Nectin-4 is a breast cancer stem cell marker that induces WNT/β-catenin signaling via Pi3k/Akt axis. Int J Biochem Cell Biol. 89:85–94. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Deng H, Shi H, Chen L, Zhou Y and Jiang J: Over-expression of Nectin-4 promotes progression of esophageal cancer and correlates with poor prognosis of the patients. Cancer Cell Int. 19:1062019. View Article : Google Scholar : PubMed/NCBI | |
|
Bellmunt J, Kim J, Reardon B, Perera-Bel J, Orsola A, Rodriguez-Vida A, Wankowicz SA, Bowden M, Barletta JA, Morote J, et al: Genomic predictors of good outcome, recurrence, or progression in high-grade T1 non-muscle-invasive bladder cancer. Cancer Res. 80:4476–4486. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Bouleftour W, Guillot A and Magne N: The anti-nectin 4: A promising tumor cells target. A systematic review. Mol Cancer Ther. 21:493–501. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
M-Rabet M, Cabaud O, Josselin E, Finetti P, Castellano R, Farina A, Agavnian-Couquiaud E, Saviane G, Collette Y, Viens P, et al: Nectin-4: A new prognostic biomarker for efficient therapeutic targeting of primary and metastatic triple-negative breast cancer. Ann Oncol. 28:769–776. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Sugiyama T, Yoneda M, Kuraishi T, Hattori S, Inoue Y, Sato H and Kai C: Measles virus selectively blind to signaling lymphocyte activation molecule as a novel oncolytic virus for breast cancer treatment. Gene Ther. 20:338–347. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Challita-Eid PM, Satpayev D, Yang P, An Z, Morrison K, Shostak Y, Raitano A, Nadell R, Liu W, Lortie DR, et al: Enfortumab vedotin antibody-drug conjugate targeting nectin-4 is a highly potent therapeutic agent in multiple preclinical cancer models. Cancer Res. 76:3003–3013. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W and Forné T: Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc. 2:1722–1733. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Schilit SLP and Morton CC: 3C-PCR: A novel proximity ligation-based approach to phase chromosomal rearrangement breakpoints with distal allelic variants. Hum Genet. 137:55–62. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Yamaguchi K, Yamaguchi R, Takahashi N, Ikenoue T, Fujii T, Shinozaki M, Tsurita G, Hata K, Niida A, Imoto S, et al: Overexpression of cohesion establishment factor DSCC1 through E2F in colorectal cancer. PLoS One. 9:e857502014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI | |
|
Kharman-Biz A, Gao H, Ghiasvand R, Zhao C, Zendehdel K and Dahlman-Wright K: Expression of activator protein-1 (AP-1) family members in breast cancer. BMC Cancer. 13:4412013. View Article : Google Scholar : PubMed/NCBI | |
|
Szalóki N, Krieger JW, Komáromi I, Tóth K and Vámosi G: Evidence for homodimerization of the c-Fos transcription factor in live cells revealed by fluorescence microscopy and computer modeling. Mol Cell Biol. 35:3785–3798. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Young DF, Andrejeva J, Li X, Inesta-Vaquera F, Dong C, Cowling VH, Goodbourn S and Randall RE: Human IFIT1 inhibits mRNA translation of rubulaviruses but not other members of the paramyxoviridae family. J Virol. 90:9446–9456. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Busse DC, Habgood-Coote D, Clare S, Brandt C, Bassano I, Kaforou M, Herberg J, Levin M, Eléouët JF, Kellam P and Tregoning JS: Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of respiratory syncytial virus. J Virol. 94:e00297–e00320. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Han Y, Bai X, Liu S, Zhu J, Zhang F, Xie L, Liu G, Jiang X, Zhang M, Huang Y, et al: XAF1 protects host against emerging RNA viruses by stabilizing IRF1-dependent antiviral immunity. J Virol. 96:e00774222022. View Article : Google Scholar : PubMed/NCBI | |
|
Haller O and Kochs G: Mx genes: Host determinants controlling influenza virus infection and trans-species transmission. Hum Genet. 139:695–705. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Kuang M, Zhao Y, Yu H, Li S, Liu T, Chen L, Chen J, Luo Y, Guo X, Wei X, et al: XAF1 promotes anti-RNA virus immune responses by regulating chromatin accessibility. Sci Adv. 9:eadg52112023. View Article : Google Scholar : PubMed/NCBI | |
|
Liao X, Xie H, Li S, Ye H, Li S, Ren K, Li Y, Xu M, Lin W, Duan X, et al: 2′, 5′-oligoadenylate synthetase 2 (OAS2) inhibits zika virus replication through activation of type I IFN signaling pathway. Viruses. 12:4182020. View Article : Google Scholar : PubMed/NCBI | |
|
Duan R, Porter W and Safe S: Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: Role of estrogen receptor Sp1 complex formation. Endocrinology. 139:1981–1990. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Binato R, Corrêa S, Panis C, Ferreira G, Petrone I, da Costa IR and Abdelhay E: NRIP1 is activated by C-JUN/C-FOS and activates the expression of PGR, ESR1 and CCND1 in luminal A breast cancer. Sci Rep. 11:211592021. View Article : Google Scholar : PubMed/NCBI | |
|
Kawahara T, Shareef HK, Aljarah AK, Ide H, Li Y, Kashiwagi E, Netto GJ, Zheng Y and Miyamoto H: ELK1 is up-regulated by androgen in bladder cancer cells and promotes tumor progression. Oncotarget. 6:29860–29876. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou H, Zarubin T, Ji Z, Min Z, Zhu W, Downey JS, Lin S and Han J: Frequency and distribution of AP-1 sites in the human genome. DNA Res. 12:139–150. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Roskoski R Jr: ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol Res. 66:105–143. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Bakiri L, Reschke MO, Gefroh HA, Idarraga MH, Polzer K, Zenz R, Schett G and Wagner EF: Functions of Fos phosphorylation in bone homeostasis, cytokine response and tumourigenesis. Oncogene. 31:1506–1517. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wang L, Yang M, Guo X, Yang Z, Liu S, Ji Y and Jin H: Estrogen-related receptor-α promotes gallbladder cancer development by enhancing the transcription of nectin-4. Cancer Sci. 111:1514–1527. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Reches A, Ophir Y, Stein N, Kol I, Isaacson B, Charpak Amikam Y, Elnekave A, Tsukerman P, Kucan Brlic P, Lenac T, et al: Nectin4 is a novel TIGIT ligand which combines checkpoint inhibition and tumor specificity. J Immunother Cancer. 8:e0002662020. View Article : Google Scholar : PubMed/NCBI | |
|
Maruoka M, Kedashiro S, Ueda Y, Mizutani K and Takai Y: Nectin-4 co-stimulates the prolactin receptor by interacting with SOCS1 and inhibiting its activity on the JAK2-STAT5a signaling pathway. J Biol Chem. 292:6895–6909. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, Callaghan K, Nicola NA, Kershaw NJ and Babon JJ: The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. 9:15582018. View Article : Google Scholar : PubMed/NCBI | |
|
Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES and Engreitz JM: Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 354:769–773. 2016. View Article : Google Scholar : PubMed/NCBI |