A comprehensive structural and functional analysis of the ligand binding domain of the nuclear receptor superfamily reveals highly conserved signaling motifs and two distinct canonical forms through evolution
- Authors:
- Published online on: January 2, 2020 https://doi.org/10.3892/wasj.2020.30
- Pages: 264-274
-
Copyright: © Mitsis et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Nuclear receptors (NRs) are one of the essential classes of transcriptional factors. NRs play a critical role in all aspects of human development, metabolism and physiology. Since they generally act as ligand-activated transcription factors, they are an essential component of cell signaling (1). Glucocorticoid receptor (GR) is part of the NRs protein superfamily that is clustered in the family of the steroid hormone. GR has been shown to interact with a variety of proteins and is a transcriptional factor that regulates multiple genes, while is simultaneously expressed in almost every cell in the human body. Glucocorticoids activate GR (2). Glucocorticoids greatly contribute to the maintenance of homeostasis and partake in the regulation of the immune system (3). They exert an impressively diverse and tissue-specific effect. In the absence of glucocorticoids, the majority of the GR resides in the cytoplasm, forming a complex with heat shock proteins. Upon ligand binding, GRs are released from the complex and translocate to the nucleus, where they influence the transcription of a number of glucocorticoid-responsive genes (2). GR is the product of a single gene, NR3C1, which was first cloned in 1985(4). NR3C1 is located on chromosome 5q31-32 in humans and undergoes alternative processing, leading to functionally distinct subtypes of GR. The human GR gene (NR3C1) consists of 9 exons. The predominant isoforms of GR in humans are hGRα and hGRβ (5). These isoforms are identical through amino acid 727, but then diverge. hGRα has an additional 50 amino acids, while hGRβ has an additional 15 non-homologous amino acids. The most well-studied isoform is hGRα, which is comprised of 777 amino acids, while hGRβ has a dominant negative effect on hGRα (6). As far as GRs taxonomy is concerned, it specifically belongs to the steroid hormone receptor (SHR) subfamily (7). GR belongs to the 3-ketosteroid receptor group, which also includes receptors for mineralocorticoids (MRs), progesterone (PR) and androgens (ARs). Another steroid hormone receptor related to the previous group, with an essential role in sexual maturation, is the estrogen receptor (ER). The majority of NRs can be considered an assembly of smaller protein modules. They share a common modular structure composed of a highly conserved DNA-binding domain (DBD; C), a less conserved ligand-binding domain/LBD (E) and some less extensively studied and highly variable N-terminal (A-B) and C-terminal domains (F). The N-terminal regions of NRs sometimes harbor potent transcriptional activation functions, known as activation function-1 (AF-1), which is independent of the LBD-ligand interaction. LBD is the site on which ligands bind, and where the main interaction with coregulators takes place. In some cases, the LBD can form the hetero- or homodimerization surfaces in NRs. Within the LBD also lies the activation function-2 (AF-2), with it referring to the recruitment of transcriptional activators in a ligand-dependent manner (8). A flexible hinge region/HR connects the DBD and LBD (D), playing a crucial role in the selection of the repertoire of DNA-binding sites. The hinge region contains a series of residues that interact with the DNA minor groove towards the C-terminal of the DBD [C-terminal extensions (CTEs)]. NRs bind to the regulatory regions of target genes as homodimers, heterodimers and monomers. The major steroid hormone receptors (GR, MR, PR, AR and ER) bind mainly, as homodimers to response elements, configured as palindromes composed of two hexad nucleotide sequences separated by 3 base pairs (9). Apart from homodimerization, monomeric binding seems to also play a significant role in tissue-specific target gene expression in GRs (10). In contrast to steroid receptors, non-steroid receptors seem to bind DNA as part of a heterodimer with retinoid X receptor (RXR). They mainly bind to response elements composed of 2 hexad half-sites arranged as tandem repeats (9). The classical mode of action of NRs implies that in the absence of their ligands, they behave as transcriptional repressors through the recruitment of specific corepressors. Ligand binding in the specific ligand binding pocket induces a conformational change of the receptor, leading to the release of the corepressors, the recruitment of co-activators and the subsequent transactivation of genes (11). A proportion of unligated NRs, and more specifically, several steroid hormones, reside in the nucleus bound to DNA (12). GR and MR are exceptions since they reside in the cytoplasm in association with a variety of proteins in the absence of ligand.
NRs form an ancient and conserved family that arose early in the metazoan lineage (11). NR molecular evolution is characterized by major events of gene duplication and gene losses. During the evolution of species, gene duplication and loss are not regularly distributed in the NR superfamily evolution (13). NR gene duplication seems to be frequent, while gene loss in the superfamily is rare. In some cases, gene loss is paralleled by duplication of a specific set of genes, which gives rise to a greater diversity of NRs. This observation suggests that the lineage-specific expansion of one gene can more than compensate for an overall trend to loss. A great difficulty emerges in studying NR evolution, since NR ligands are distinctive. NR ligands are small molecules not encoded by genes; hence, the ligand is not the product of a single gene, but the product of a metabolic pathway which interacts with other pathways. This attribute is an important facet of their role in signaling. Therefore, evolution does not occur only on a single gene by the slow accumulation of mutations, but on an entire network of genes (11). Thus, predicting how the specificity of a receptor for its ligand evolves is not an easy task.
Data on the origins of NRs suggest that they were not a hormonal receptor with high affinity for a particular ligand and that feature was acquired later during evolution. It is considered that the first NR had the ability to bind different molecules with small affinity (14). This is not surprising, since data suggest that a single NR can bind several ligands, with different biological activities, and those ligands can act as modulators that can selectively activate an NR in a tissue or target specific manner (11). Consequently, investigating the mechanisms through which selectivity functions in NRs is an important step in understanding their evolutionary history. An analysis of the mechanisms through which mutations alter the selectivity and function of receptors can be considered a valid method in decrypting NR receptor evolution.
Data and methods
Dataset collection and filtering
A search was conducted on the RSCB Protein Data Bank (PDB) database (15) for amino acid sequences that are related to the NR LBD, followed by the acquisition of resulting data. Specific sequences that responded to the query but did not include the LBD were removed from the dataset, by using regular expressions techniques and local alignments with reference sequences. In total, 420 NR ligand binding domain protein structures were downloaded from several species.
Sequence alignment
Multiple sequence alignment was performed using the MATLAB Bioinformatics Toolbox, utilizing the progressive multiple alignment method and a guide tree (16,17). Pairwise distances between protein sequences were computed following pairwise alignment with the Gonnet scoring matrix (18) and followed by counting the proportion of sites at which each pair of sequences are different. The guide tree was calculated by the neighbor-joining method assuming equal variance and independence of evolutionary distance estimates. The visualization of the consensus sequence was performed using the JalView platform (19) and based on the multiple sequences alignment results and parameters such as amino acids quality and conservation. A more specific alignment with representative members of the Steroid hormone receptors was performed using the Molecular Operating Environment (MOE) (20).
Mutation analysis
A collection of currently known natural occurring NR LBD mutations was assembled following a literature review. The selected mutations harmed ligand binding. The resulting dataset includes natural occurring LBD mutations in the following receptors: GR, MR, AR, ERα, peroxisome proliferator-activated receptor (PPAR)γ, retinoid acid receptor (RAR)α, RARβ, thyroid hormone receptor (THR)α, THRβ, liver X receptor (LXR)α, vitamin D receptor (VDR), hepatocyte nuclear factor 4 (HNF4A), steroidogenic factor 1 (SF1), RAR-related orphan receptor (ROR)α, RORβ and RORγ (Table SI). Based on the generated list, all mutations were detected and marked on the consensus sequence from the multiple sequence alignment, and in the alignment of the representative structures of the SHR. Additionally, the mutation rate was examined in all NRs (Table SII). Specific mutation positions were highlighted by different colors in order to showcase the mutation rate within the NRs (Fig. S1).
Structural and functional analysis
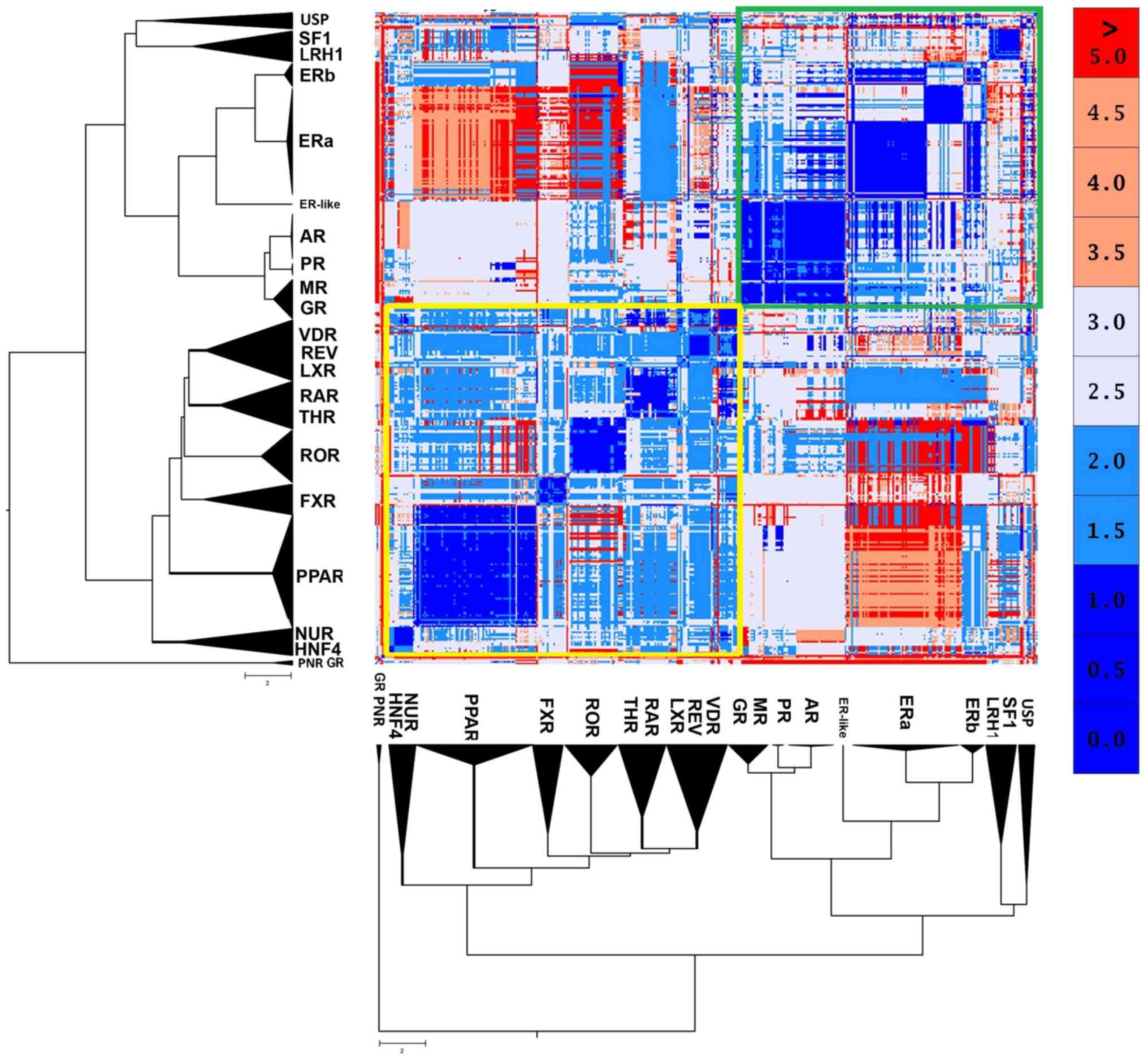
A thorough analysis of NR LBD structural comparisons was achieved by superimposing the structures and by calculating a matrix of root mean square deviation (RMSD). The structural comparisons between the 420 structures were performed utilizing the structural superposition method, as described in the MATLAB Bioinformatics Toolbox (21). This method computes and applies a linear transformation to superpose the coordinates of the atoms of the first structure to the coordinates of the atoms of the second structure. Alpha carbon atom coordinates of single chains for each structure are considered for computing the linear transformation. The structural similarity matrix was displayed using MATLAB in 5 different colors (blue for the range 0 to 1, light blue for the range 1.1 to 2, light gray for the range 2.1 to 3, orange for the range 3.1 to 4.9 and red for the values ≥5).
A more in-depth look at steroid hormone receptor structures was gained using MOE (20,22-24), where all the extracted mutations and interactions sites with several proteins and ligands were identified and studied. Specifically, each PDB entry was examined for ligand interaction using the ligand interaction function. MOE showcased the LBD amino-acids that interacted with said ligands or co-activators. Finally, beneficial information from the NCBI conserved domain database was extracted and assigned with the MOE results in the consensus sequence from the multiple sequences alignment.
Phylogenetic analysis
A specialized phylogenetic analysis was performed using the MATLAB Bioinformatics Toolbox (20) utilizing the Unweighted Pair-Group Method (UPGMA) (25-27) and a specific hybrid matrix of pairwise distances (28). This matrix combines both information from the distance matrix of the multiple sequence alignment, and the RMSD matrix of the structural analysis. The combined specialized matrix is calculated through element by element matrices proliferation (29,30). This technique allows the clustering of less similar proteins in sequence level that are more conserved in the structural level. Finally, the constructed phylogenetic tree was visualized using the MEGA (31) radiation option, and the final clusters were separated by different colors.
Ligand analysis
The notion of chemical similarity plays an important role in predicting the properties of chemical compounds, clustering chemicals, and in particular, in conducting functional analysis studies. The calculation of the similarity of any two molecules is achieved by comparing their molecular fingerprints (32). These fingerprints are comprised of structural information about the molecule which has been encoded as a series of bits. The most popular similarity measure for comparing chemical structures represented by means of fingerprints is the Tanimoto coefficient (33).
First, a list with all extract ligands that co-crystallized in the corresponding SHR LBD structures was created (Table SIII). We set the same order in the list as the structures' order from the phylogenetic analysis. The structural comparisons between the 179 SHR ligands were performed using the Tanimoto coefficient algorithm (34). The Tanimoto coefficient ranges from 0, when the fingerprints have no bits in common, to 1, when the fingerprints are identical. In the end, all the similarities were saved in a chemical-specific similarity matrix. Finally, the chemical-specific similarity matrix was displayed using MATLAB in 4 different colors (blue for the range 1 to 0.9, light blue for the range 0.89 to 0.7, purple for the range 0.69 to 0.6 and black for the range 0.59 to 0).
Results
The 4 major clusters of the NR LBD
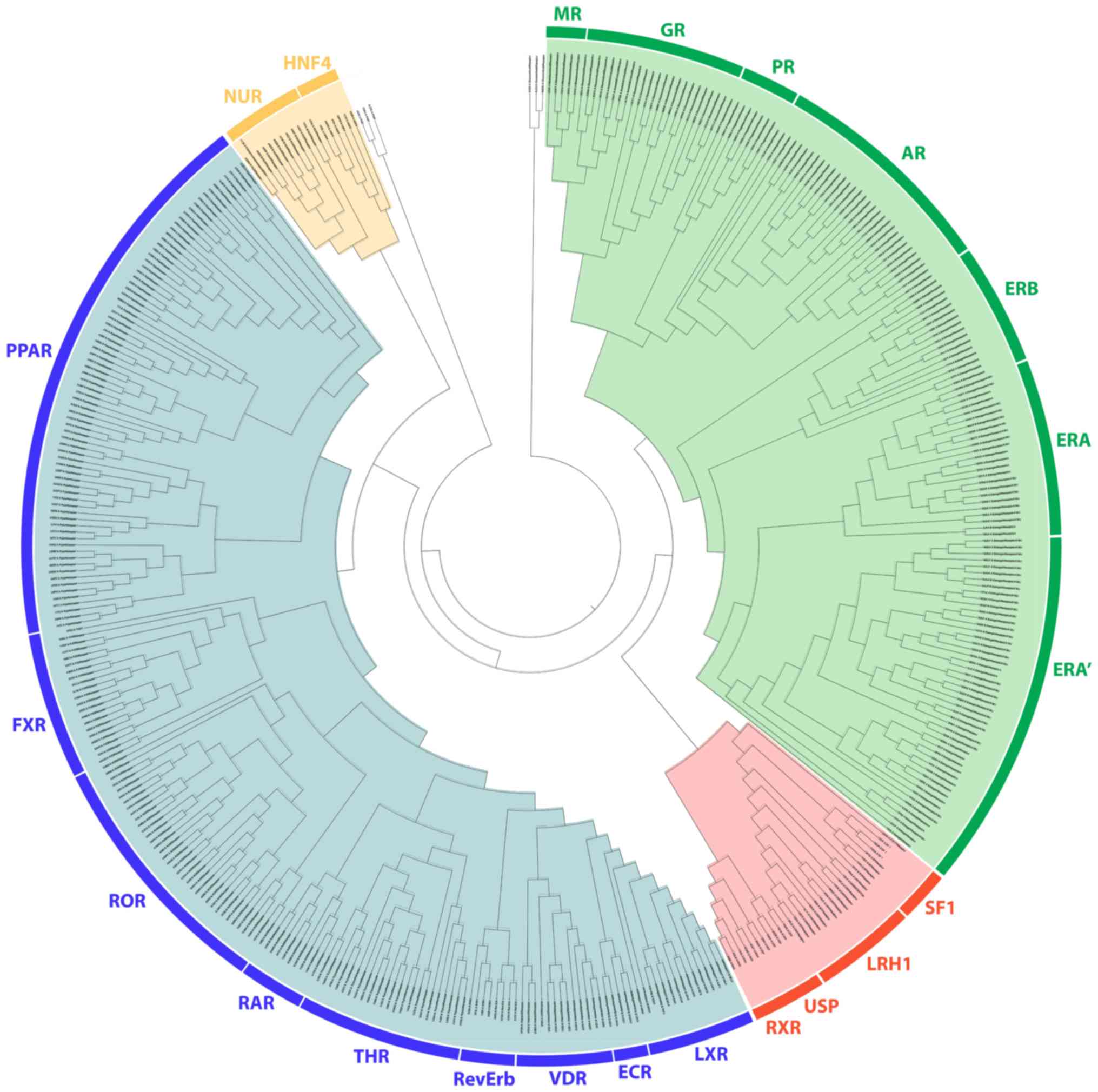
Phylogenetic analysis revealed a distinct separation of NR LBDs into 4 monophyletic branches, the steroid hormone receptor-like cluster, the thyroid hormone-like receptors cluster, the retinoid X-like and steroidogenic factor-like receptor cluster and the nerve growth factor-like/HNF4 receptor cluster (Fig. 1). All known steroid hormone receptors (GRs, MRs, PRs, ARs and ERs) have been grouped in separated sub-clusters in the steroid hormone-like receptor cluster. As expected, the LBD of GRs, MRs, PRs and Ars was found to be well-related to the ERs. The ER receptors also separate into two different subclasses, the ERα and the ERβ, from which there is a substantial separation of the ERα in 2 groups. A more detailed structural analysis was performed in order to understand this strange distribution of the ERs, and the results are described below. Moreover, the SRH-like cluster was found to be directly related with the second cluster of the retinoid X-like and steroidogenic factor-like receptor cluster. The linkage of this protein family with the steroid hormone receptor family was noted for the first time, to the best of our knowledge. Within the second cluster are contained the RXR, the liver receptor homolog 1 (LRH1), the SF1 and the ultraspiracle protein (USP) subunit of the ecdysone receptor. The SF1 is a protein that controls many aspects of adrenal and reproductive function. It is encoded by the NR5A1 gene, which is a member of the NR protein family (35). Along with the LRH1 encoded by the NR5A1 gene, they belong to the steroidogenic factor-like subfamily of NRs. They both hold a critical role in steroidogenesis (36). Another thoroughly different observation on the SHR branch is its inclusion of the RXR and its Drosophila homolog USP, which is a subunit of the ecdysone receptor (37,38).
On the third monophyletic branch, the LBD of the THR-like cluster, all known members are grouped, including THR, PPAR, LXR, VDR, RAR, FXR, ROR and orphan nuclear hormone receptor (NR1D1; RevErb). The EcR subunit of the ecdysone receptor also belongs in this cluster. The EcR subunit of the ecdysone receptor is closely related to the FXR, based on their similarities in the DNA binding domain (39). The EcR/USP heterodimer seems to be the corresponding arthropod complex to the FXR/RXR heterodimer. This analysis provides the insight that this heterodimer appears to be composed of 2 subunits with each one belonging to a different monophyletic branch. Moreover, the LBD of the THR-like cluster was found to be directly related to the fourth cluster of the LBDs of the nerve growth factor (Nurr77) and HNF4. HFN4α and Nurr77 are suggested to be in the same subfamily. Some other interesting observations are the existence of a number of distinct GRs showcasing marked differences with regard to other NRs.
The conserved signaling motifs of the NR LBD
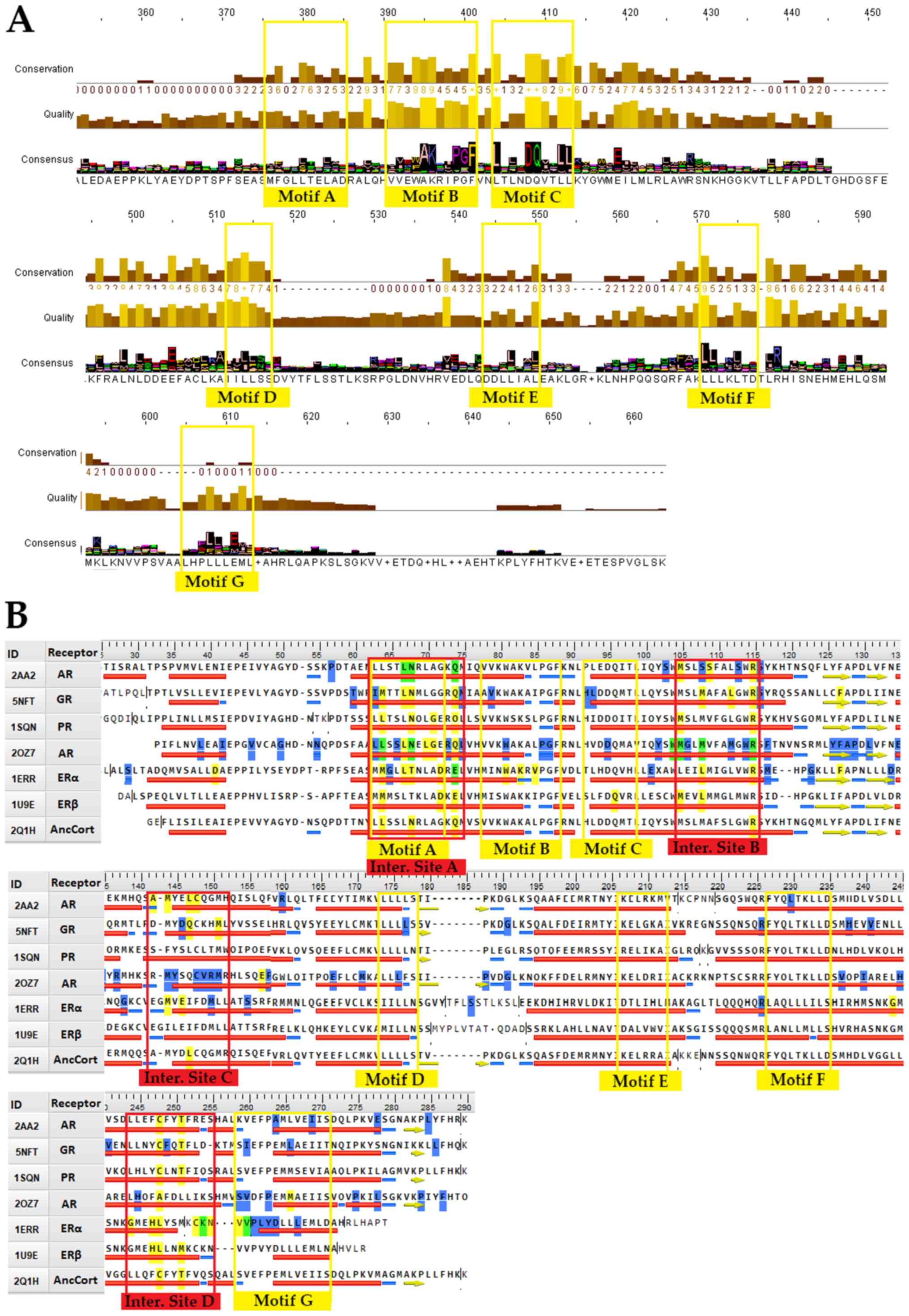
An in-depth review of the consensus sequence and NR LBD mutations provides a basis for 7 highly conserved signaling motifs (Fig. 2A). A more targeted approach to steroid hormone receptors can provide data on their importance for NR function (Fig. 2B). Motif A occupies consensus sequence positions 378 to 385 (Fig. 2A). This LLxxL motif is an inverse NR-box (LLxxL) and still contains the ability to interact with several NRs (40). Based on NCBI's conserved domain database (CDD) (41), this position is one of the main ligand interaction sites of the family and is of utmost importance to NRs. It also coincides with the SHR interaction Motif A (Fig. 2B). Motif B occupies consensus sequence positions 391 to 401 (Fig. 2A). A cross-check with CCD NR action sites (Fig. 2B) reveals that Motif B resides in a region critical to co-activator function. The GR mutation V575G does support this hypothesis (42). V575 is part of the AF-2 surface, and it contributes directly to the attraction of LxxLL (NR-box sequence motif). A mutation on that specific region hinders the ability of co-activators to interact with the NR. The 1L2I PDB entry for ERα also implies the region's high impact role in interacting with co-activators, and more specifically, the NR-box sequence motif. Motif C occupies positions 404 to 413, a region that is also critical to co-activator function (Fig. 2A). Motif C could be described as LxxDDQ. Along with the R (402 alignment position) residue, this motif creates a structure specific to GRs, ARs and PRs. The study by Bledsoe et al was the first one that recognized this specific region's critical role in LBD function (7). Motif C partakes in the creation of GR's second charge clamp. This distinct structure is vital to the binding of the third NR-box of co-activators. The residues responsible for the second charge clamp are not present in the remaining SHRs (ERs and MRs). Motif D occupies positions 512 to 516. This region is part of the highly conserved C-terminal end of the helix eight domain in SHRs, whose function is critical to ligand binding. Mutations in this motif, including GR's L672P and AR's L813P lead to a complete lack of ligand binding (43,44). More experiments also showcase that the mutant proteins are possibly prone to higher degradation (44). Motif E is an inverse NR-box and occupies positions 546 to 550. Motif F occupies positions 568 to 575. This LxxLL motif (NR-box) is found in all SHRs, with the ERα LxxLL comprising positions 568 to 572. Motif G occupies alignment positions 601-613. It contains an ERα LxxLL motif on positions 601 to 609, a PPAR LxxLL motif on positions 605 to 609, and an LLxxL ERα motif on position 609 to 611. It is quite intriguing that ERα exhibits a motif of LxxLLLxxL, which contains both an NR-box and its inverse. A prime example of a malfunctioning mutation on motif G is the Y537S, which is present on ERαs appearing in breast cancer cells (45).
Mutations in the NR LBD
A list of all known NR LBD natural mutations was composed in this study from previous publications (Tables SI and SII). Studying both the common natural mutations and their results gives way to a few observations. Firstly, highly conserved sequences are not prone to mutations (Fig. S1). They seem critical to normal protein function, which leads to their evolutionary conservation. Indeed, natural mutations on highly conserved positions lead to deliberating effects. Some positions exhibit no mutation at all. The lack of mutations may be attributed to their quintessential role in protein function, since mutations on said positions could be lethal, so no surviving phenotypes exist. The majority of mutations on steroid receptors are associated with hormone level changes, which are linked to their respective ligand. It should be mentioned that phenotypes that could be attributed to mutations on NRs may not exhibit said mutations on the final protein product. A number of factors could be responsible for this observation, such as epigenetic actions on NR genes, mutations in non-coding regions affecting enhancers, or mutations on NR cofactors (46). An in-depth look into GR mutations confirmed the aforementioned. No mutations were found on highly conserved NR amino acids. Mutations on GR LBD have a severe effect on adrenocortical function. Mutations of GR LBD can lead to Chrousos syndrome, a condition characterized by tissue insensitivity to glucocorticoids. An interesting fact is that some LBD mutations have a dominant negative effect. These kinds of mutations on the LBD can be considered more severe than DBD mutations since they affect normal functioning proteins as well.
The 2 major canonical forms
NRs are structurally quite conserved. However, the structural analysis results of NR LDB displayed 2 distinct canonical forms (Fig. 3). The first one seems to be prevalent in the SHR-like, while the second one is prevailing in the THR-like receptors. The well-defined retinoid X receptor-like/steroidogenic factor-like cluster of the USP, SF1 and LRH1 receptors may appear phylogenetically close, although they have a small structural correlation with the two major forms. Based on these findings, the SHR-like LBD canonical form exhibits highly conserved structural domains, albeit it validates the notion above, that estrogen receptors are pretty different from the rest of SHRs. A peculiar observation emerges, one in which ERβ found to be more structurally similar to the rest of SHRs than ERα (Fig. 3). Another observation is the number of β-strands that compose SHRs is 4, while the number of α-helixes is not constant in each specific entry. Based on crystallographic observations, a steroid hormone receptor is formed either with 11 or 12 α-helixes. A specific examination in an alignment featuring only GRs also showcases the different activation states of NRs, based on the position of the AF-2 containing helix (7). The activation state is dependent on the ligand featured and the presence or absence of co-activators/corepressors. On the other hand, 3 GR LBD structures (PDB: 3H52, 4LSJ and 4MDD) and 3 photoreceptor-specific NRs (PNR) LBD structures (PDB: 4LOG, 4XAJ and 4XAI) appear to distance themselves from the entirety of all NR LBD structures. An in-depth look proposes that those GR LBD structures correspond to a specific antagonist form of the receptor, in which the dislocation of the 12th helix leads to complete disruption of the receptor's function (21). The second major canonical form of the thyroid hormone-like receptor LBD is highly conserved, with small differences amongst receptors. These differences create somewhat distinct subclasses, the PPAR-like, the ROR/THR, such as VDR-like and the HNF4/Nur77-like.
The 3 ligand specificity clusters of the SHR LBD
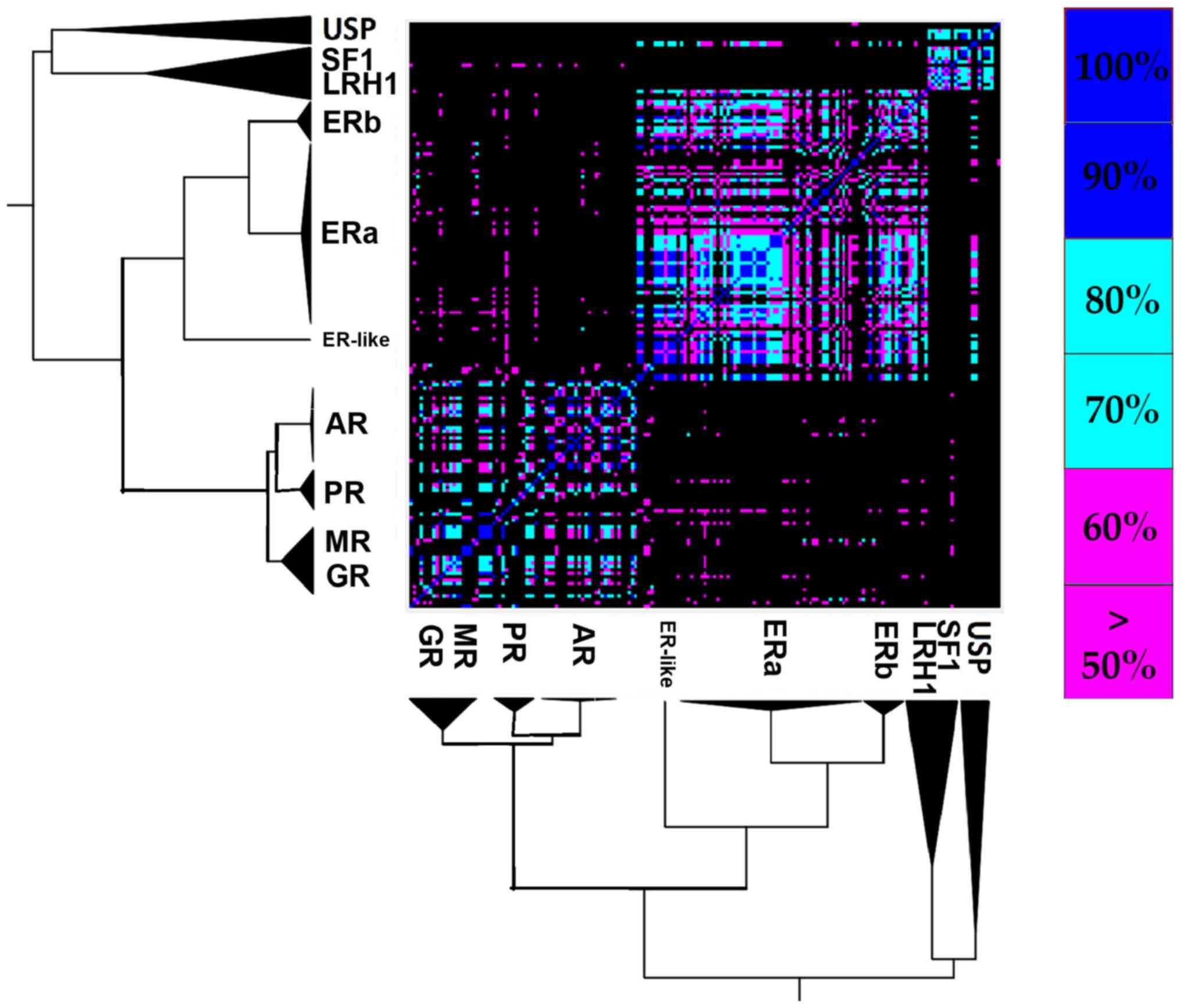
Ligand specificity is one of the major and most important characteristics of NRs (47). NR ligands are small hydrophobic molecules that bind their corresponding NR LBD's hydrophobic pocket. A list featuring all SHR ligands that were co-crystallized in the corresponding SHR structures was created (Table SIII). The majority of ligands seem to be receptor-specific, with the exceptions of MOF, which binds both GR and PR and R18, which binds both PR and AR. This observation is quite interesting since, as mentioned above, AR, MR, PR and GR appear to create a steroid receptor sub-class of their own, different from ERs. The PDB entry 1GS4 sheds light onto the specific association between GR, PR and AR (48). Mutation T877A on the AR adds the ability to bind progesterone 17b-estradiol and some anti-androgens. The threonine at position 877 on AR is unique, though its corresponding alignment position (251, Fig. 2B) partakes in ligand interactions in all of the steroid receptors. This mutation gives the AR abilities that have more in common with PR and ER. Mutation L701H substantially impairs AR's own ability to bind androgen, but allows it to bind cortisol (48). The leucine at the 701 position is present in MR, PR and AR, while its corresponding alignment position (67, Fig. 2B) partakes in ligand interaction in GRs, PRs, ARs and ERs.
A comparative analysis of the ligands that have been co-crystallized with their corresponding receptors was conducted, in order to create a more clear-cut idea for the interactions that are characteristic of the steroid hormone receptors' ligand binding pocket (Fig. 4). A total of 94 unique steroid hormone receptors ligands were collected and were compared in order to search for similarities and identify new relations. Based on the results, there is a clear separation in the main clusters, as shown in Fig. 4. Those are the USP/SF1/LRH1 ligand-specific cluster, the ER ligand-specific cluster and the AR/PR/ MR/ GR ligand-specific cluster. It is quite interesting that the USP/SF1/LRH1 ligand-specific cluster contains ligands that are similar to the ER ligand-specific cluster. Moreover, focusing on the ER ligand-specific cluster, there is a clear separation of ERα in 2 sub-clusters.
A sub-cluster of ERas forms and acts as estrogen β, and the risk of breast cancer
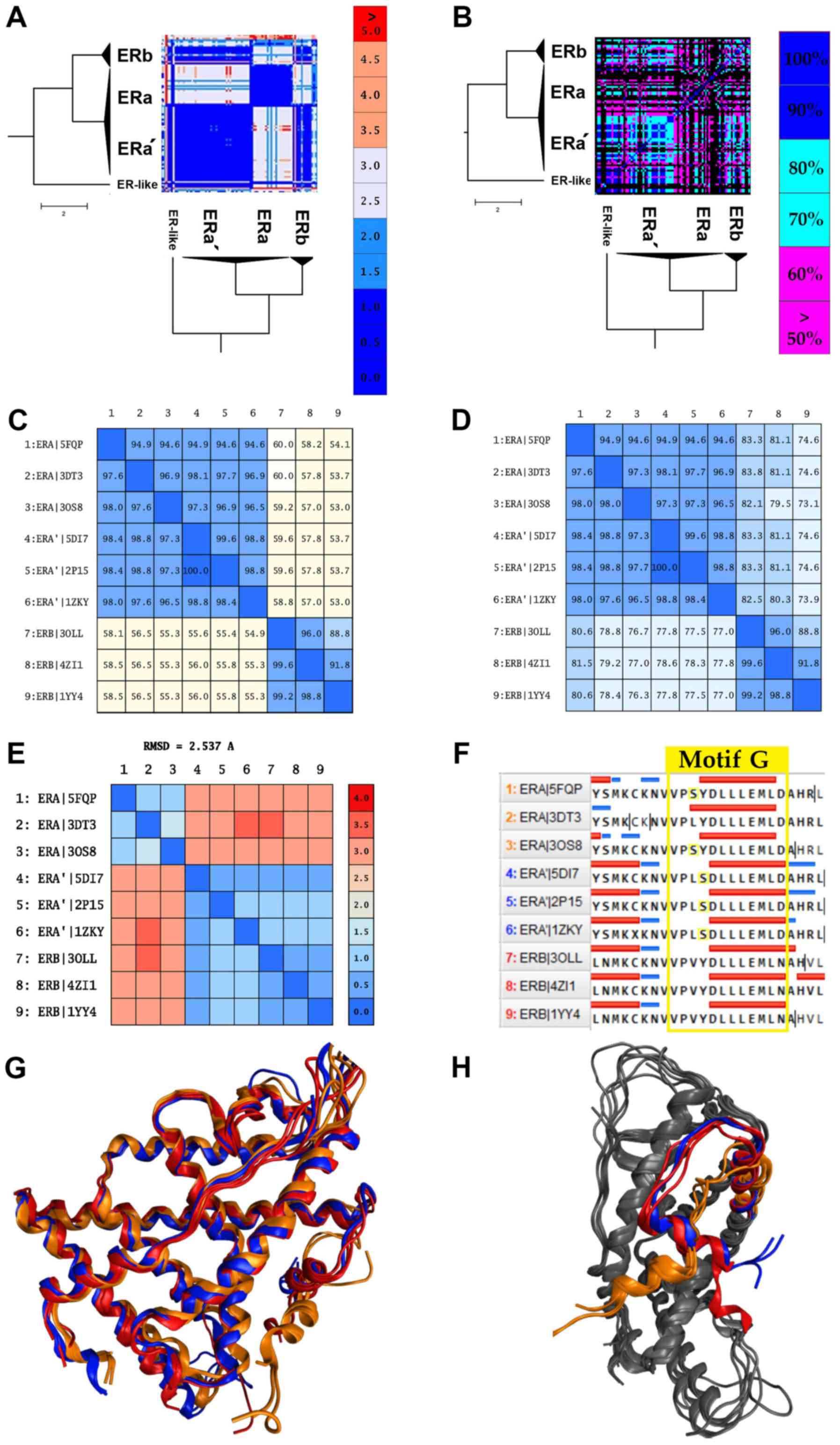
As expected, the majority of NRs in the same subfamily display high structural similarity, with a great example being the ARs, PRs and MRs (Fig. 3). A review of the SHR-like branch adds some new information. Contrary to the observations, some receptors indicate significant structural differences even among themselves (Fig. 3). This observation points out that the structures within the same category share more than one canonical form due to a critical mutation, disease, or cancer. The ERα data are provided in the same direction. Despite ERα showing high sequence similarity (Fig. 5D), it is structurally separated into 2 different canonical forms (Fig. 5A). An in-depth look at the ERα structures comprising of the 2 canonical forms yielded some interesting results. The 2 canonical forms of ERα correspond to 2 different types of ERα.
The first canonical form of ERα contains various mutant ERαs and a small number of wild-type receptors. The most common mutants in this sub-cluster are located at positions C381, C417, C530 and L536. The second canonical form of ERα, which will be referred to as ERα', is mainly characterized by the Y537S mutation. This mutation is commonly found in breast cancer patients and is associated with resistance to several endocrine therapies (49). More specifically, this mutation is located in the AF-2 helix of the ERα' LBD and shifts the receptor equilibrium towards the agonist conformation, even in the absence of a ligand (23). Moreover, the ERα' canonical form appears to be identical in the structural level with the ERβ (RMSD <2) rather than the ERα, while in the sequence level, all ERαs and ERαs' exhibit minimal differences (Fig. 5A, C, E and G). Based on these findings, the Y537S mutation induces a critical conformational change in the ERα structure. In particular, it changes the angle of the AF-2 helix of the ERα' LBD, in a position which is identified coequally on the ERβ AF-2 helix (Fig. 5H). The displacement and the 90˚ turn of the AF-2 helix plays a key role in the action of the ERα' since it contains the signaling ‘Motif G’, one of the most important motifs of the NR LBD (Fig. 5F). This hypothesis seems to be confirmed with the functional analysis we had performed by analyzing all the available ligands and chemicals that have been co-crystallized in the cavity of the ERα' and generally in all ERs. Based on these results, ERα' can interact without any specificity with all the identified ligands on ERs, including ERβ (Fig. 5B). This particularity has also been described by several studies (48-50) that are found to refer to a number of ERα' structures. In the study by Nettles et al, a member of the ERα' (PDB: 2P15) was crystallized, and it was concluded that this ERα could interact with a wider array of pharmacophores than was previously thought (50). In summary, it is possible that the Y537S mutation induces a LBD domain with higher plasticity in the ERα'.
Discussion
Hybrid phylogenetic analysis is more eloquent than a common phylogenetic analysis. It combines both sequence and structural information from the NRs' LBD to propose clusters with higher confidence. It has been observed that protein structure is more conserved than sequence (51), something visible on the current analysis. Differences that are not detected on sequence analysis alone are visible on this analysis. The phylogenetic analysis performed depicts the distinct separation of NR LBDs into the monophyletic branches of the steroid hormone receptor-like cluster, the thyroid hormone-like receptors cluster, the retinoid X-like and steroidogenic factor-like receptors cluster, and the nerve growth factor-like/HNF4 receptors cluster and the LBD of GRs, MRs, PRs and ARs found to be well-related to ERs. The linkage of the SRH-like cluster with the second cluster of the retinoid X-like and steroidogenic factor-like receptors cluster is noted for the first time, at least to the best of our knowledge.
Researching motifs in the NR ligand binding domain is of utmost importance. As mentioned above in the ‘Introduction’, the ligand binding domain is a somewhat conserved domain. Finding motifs that have been conserved through the evolutionary process should highlight regions of critical importance in ligand binding. Those regions can provide amino acid patterns that are a unique signature to the NR LBD. Indeed, a previous study published in 1996 found signature sequences that are essential in maintaining the ligand binding pocket structure (52). Those sequences seem to agree with the proposed motifs that are described below, and some new ‘key’ motifs are introduced. The existence of NR-boxes or inverse NR-boxes on SHRs may seem peculiar on first glance, but it should not be a surprise. NRs have the ability of both hetero- and homo-dimerization. Since NR-boxes can bind to specific NR regions, they may have a role in the interaction between NRs, specifically hetero and homo-dimerization. The results showcase moderate conservation of specific amino acid sequences. Length-wise all NR ligand binding domains are pretty similar. Looking through the subcategory of steroid hormone receptors also provides some interesting insights. There are many more sequence similarities, as expected, though it is interesting, that estrogen receptors seem to create a subcategory of their own since they exhibit amino acid variations not present in the rest steroid hormone receptors.
Steroid hormone receptors share 4 main interaction sites from where directly interact with several ligands and co-activators (Fig. 2B). The interaction sites A and B of the SHR LBD are well characterized and found across all NR members. The interaction sites C and D should be highly variable among NRs and somewhat conserved on SHRs. The incorporation of both mutations and ligand interaction points can specify the importance of these action regions. Fig. 2B provides information on both interaction points and mutations. It shows that all interaction sites are prone to mutations. This fact also seems logical, since the activation sites are the ones responsible for the selectivity of NRs. Mutations in those regions possibly are at the forefront of NR evolution. The consensus sequence also adds to this speculation, since the main interaction regions are characterized by relatively small sequence conservation (Fig. 2B). It should also be highlighted that the interaction sites A and B, which are conserved on all NRs, seem to be bridged by 3 regions of highly conserved motifs A, B and C.
Based on the comparative analysis of the co-crystallized ligands to their receptors, there is a clear separation in the main clusters, the USP/SF1/LRH1 ligand specific cluster, the ER ligand specific cluster, and the AR/PR/MR/GR ligand specific cluster. The USP/SF1/LRH1 ligand specific cluster contains ligands similar to the ERs ligands specific cluster, where there is evident separation of ERα in two sub-clusters. Moreover, ERα' can interact without any specificity with all the identified ligands on ERs, including ERβ, and there is a possible evidence that the Y537S mutation induces a LBD domain with higher plasticity in the ERα'.
In conclusion, NRs are vital transcription factors, and the aforementioned information acquired can be used in a variety of ways. The interaction sites and conserved motifs can be used as selected targeted regions for novel drugs. The development of new drugs can be achieved through specific in silico techniques that can compose ligands, which can interact with those specific regions and force new alterations in the protein's dynamics (53). The phylogenetic analysis also provided new insights for NR clustering and identified several key regions that exist through evolution in the NR LBD such as the amino acid repeating motif ‘LxxLL’ or ‘LLxxL’. The mutation analysis highlighted mutational hotspots, while also providing insights on their effects in structure and function, especially when they are localized in NR conserved signaling motifs. Structural and functional analysis of the NR LBD display two major canonical forms and identify 3 ligand specific clusters within the steroid hormone receptor family. Last but not least, a new sub-cluster of ERα with a very specific canonical form has been identified and related to breast cancer through a well-known mutation in ERs. This new information may be of high importance in order to understand the signaling mechanism underlying NRs and cancer.
Supplementary Material
Mutation rates on the consensus sequence from the multiple sequence alignment. Specific mutation positions are highlighted by different colors in order to showcase frequencies. Blue is used to mark positions which showcase mutations on two different NRs, and green is used to mark positions which showcase mutations on 3 different NRs; red is used to mark positions which showcase mutations on four different NRs, while purple is used to mark positions which showcase mutations on 5 different NRs. NRs, nuclear receptors.
List of various nuclear receptor mutations located in the ligand binding domain.
Mutations rates on different nuclear receptors based on the multiple alignment position.
List featuring all SHR ligands and their corresponding receptors.
Acknowledgements
Not applicable.
Funding
DV would like to acknowledge funding from: i) Microsoft Azure for Genomics Research Grant (CRM:0740983); ii) FrailSafe Project (H2020-PHC-21-2015-690140) ‘Sensing and predictive treatment of frailty and associated co-morbidities using advanced personalized models and advanced interventions’, co-funded by the European Commission under the Horizon 2020 research and innovation program; iii) Amazon Web Services Cloud for Genomics Research Grant (309211522729); iv) AdjustEBOVGP-Dx (RIA2018EF-2081): Biochemical Adjustments of native EBOV Glycoprotein in Patient Sample to Unmask target Epitopes for Rapid Diagnostic Testing. A European and Developing Countries Clinical Trials Partnership (EDCTP2) under the Horizon 2020 ‘Research and Innovation Actions’ DESCA. EE would like to acknowledge funding by the project ‘INSPIRED-The National Research Infrastructures on Integrated Structural Biology, Drug Screening Efforts and Drug Target Functional Characterization’ (Grant MIS 5002550) and by the project: ‘OPENSCREEN-GR An Open-Access Research Infrastructure of Chemical Biology and Target-Based Screening Technologies for Human and Animal Health, Agriculture and the Environment’ (Grant MIS 5002691), which are implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Programme ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
TM, LP, AE, FB, DV, GPC, EE have all equally contributed to the writing, drafting, revising, editing, reviewing, and the conception and design of the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Robinson-Rechavi M, Escriva Garcia H and Laudet V: The nuclear receptor superfamily. J Cell Sci. 116:585–586. 2003. View Article : Google Scholar | |
|
Chrousos GP: The glucocorticoid receptor gene, longevity, and the complex disorders of Western societies. Am J Med. 117:204–207. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Bereshchenko O, Migliorati G, Bruscoli S and Riccardi C: Glucocorticoid-Induced Leucine Zipper: A Novel Anti-inflammatory Molecule. Front Pharmacol. 10(308)2019.PubMed/NCBI View Article : Google Scholar | |
|
Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG and Evans RM: Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 318:635–641. 1985.PubMed/NCBI View Article : Google Scholar | |
|
Nicolaides NC, Galata Z, Kino T, Chrousos GP and Charmandari E: The human glucocorticoid receptor: Molecular basis of biologic function. Steroids. 75:1–12. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Kadmiel M and Cidlowski JA: Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 34:518–530. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, et al: Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 110:93–105. 2002.PubMed/NCBI View Article : Google Scholar | |
|
Huang P, Chandra V and Rastinejad F: Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu Rev Physiol. 72:247–272. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Evans RM and Mangelsdorf DJ: Nuclear Receptors, RXR, and the Big Bang. Cell. 157:255–266. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Lim HW, Uhlenhaut NH, Rauch A, Weiner J, Hübner S, Hübner N, Won KJ, Lazar MA, Tuckermann J and Steger DJ: Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 25:836–844. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Holzer G, Markov GV and Laudet V: Evolution of Nuclear Receptors and Ligand Signaling: Toward a Soft Key-Lock Model? Curr Top Dev Biol. 125:1–38. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Klinge CM: Steroid Hormone Receptors and Signal Transduction Processes. In: Principles of Endocrinology and Hormone Action. Belfiore A and LeRoith D (eds). Springer International Publishing, Cham, pp187-232, 2018. | |
|
Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V and Robinson-Rechavi M: Evolutionary genomics of nuclear receptors: From twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 21:1923–1937. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Markov GV and Laudet V: Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol. 334:21–30. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The Protein Data Bank. Nucleic Acids Res. 28:235–242. 2000.PubMed/NCBI View Article : Google Scholar | |
|
Sobie EA: An introduction to MATLAB. Sci Signal. 4(tr7)2011.PubMed/NCBI View Article : Google Scholar | |
|
Papageorgiou L, Loukatou S, Sofia K, Maroulis D and Vlachakis D: An updated evolutionary study of Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase reveals novel invariable motifs as potential pharmacological targets. Mol Biosyst. 12:2080–2093. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Pearson WR: Selecting the Right Similarity-Scoring Matrix. Curr Protoc Bioinformatics. 43:3.5.1–3.5.9. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Waterhouse AM, Procter JB, Martin DM, Clamp M and Barton GJ: Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Vilar S, Cozza G and Moro S: Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 8:1555–1572. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Kufareva I and Abagyan R: Methods of protein structure comparison. Methods Mol Biol. 857:231–257. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Aertgeerts K, Skene R, Yano J, Sang BC, Zou H, Snell G, Jennings A, Iwamoto K, Habuka N, Hirokawa A, et al: Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J Biol Chem. 286:18756–18765. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Papageorgiou L, Loukatou S, Koumandou VL, Makałowski W, Megalooikonomou V, Vlachakis D and Kossida S: Structural models for the design of novel antiviral agents against Greek Goat Encephalitis. PeerJ. 2(e664)2014.PubMed/NCBI View Article : Google Scholar | |
|
Papageorgiou L, Megalooikonomou V and Vlachakis D: Genetic and structural study of DNA-directed RNA polymerase II of Trypanosoma brucei, towards the designing of novel antiparasitic agents. PeerJ. 5(e3061)2017.PubMed/NCBI View Article : Google Scholar | |
|
Michener CD and Sokal RR: A quantitative approach to a problem in classification. Evolution. 11:130–162. 1957. View Article : Google Scholar | |
|
Sneath PHA and Sokal RR: Unweighted pair group method with arithmetic mean. In: Numerical Taxonomy. W.H. Freeman, San Francisco, CA, pp230-234, 1973. | |
|
Pavlopoulos GA, Soldatos TG, Barbosa-Silva A and Schneider R: A reference guide for tree analysis and visualization. BioData Min. 3(1)2010.PubMed/NCBI View Article : Google Scholar | |
|
Lu J, Xu G, Zhang S and Lu B: An effective sequence-alignment-free superpositioning of pairwise or multiple structures with missing data. Algorithms Mol Biol. 11(18)2016.PubMed/NCBI View Article : Google Scholar | |
|
Leaché AD, Wagner P, Linkem CW, Böhme W, Papenfuss TJ, Chong RA, Lavin BR, Bauer AM, Nielsen SV, Greenbaum E, et al: A hybrid phylogenetic-phylogenomic approach for species tree estimation in African Agama lizards with applications to biogeography, character evolution, and diversification. Mol Phylogenet Evol. 79:215–230. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Fouquier J, Rideout JR, Bolyen E, Chase J, Shiffer A, McDonald D, Knight R, Caporaso JG and Kelley ST: Ghost-tree: Creating hybrid-gene phylogenetic trees for diversity analyses. Microbiome. 4(11)2016.PubMed/NCBI View Article : Google Scholar | |
|
Stecher G, Liu L, Sanderford M, Peterson D, Tamura K and Kumar S: MEGA-MD: Molecular evolutionary genetics analysis software with mutational diagnosis of amino acid variation. Bioinformatics. 30:1305–1307. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Mellor CL, Marchese Robinson RL, Benigni R, Ebbrell D, Enoch SJ, Firman JW, Madden JC, Pawar G, Yang C and Cronin MTD: Molecular fingerprint-derived similarity measures for toxicological read-across: Recommendations for optimal use. Regul Toxicol Pharmacol. 101:121–134. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Rácz A, Bajusz D and Héberger K: Life beyond the Tanimoto coefficient: Similarity measures for interaction fingerprints. J Cheminform. 10(48)2018.PubMed/NCBI View Article : Google Scholar | |
|
Bajusz D, Rácz A and Héberger K: Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminform. 7(20)2015.PubMed/NCBI View Article : Google Scholar | |
|
Ferraz-de-Souza B, Lin L and Achermann JC: Steroidogenic factor-1 (SF-1, NR5A1) and human disease. Mol Cell Endocrinol. 336:198–205. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Fayard E, Auwerx J and Schoonjans K: LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14:250–260. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Hall BL and Thummel CS: The RXR homolog ultraspiracle is an essential component of the Drosophila ecdysone receptor. Development. 125:4709–4717. 1998.PubMed/NCBI | |
|
Hill RJ, Billas IM, Bonneton F, Graham LD and Lawrence MC: Ecdysone receptors: From the Ashburner model to structural biology. Annu Rev Entomol. 58:251–271. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD and Edwards PA: Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 275:10638–10647. 2000.PubMed/NCBI View Article : Google Scholar | |
|
Greschik H, Flaig R and Moras D: Ligand/cofactor complexes of nuclear receptor ligand-binding domains. https://www.academia.edu/18571843/Ligand_cofactor_complexes_of_nuclear_receptor_ligand-binding_domains. | |
|
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al: CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Nicolaides NC, Roberts ML, Kino T, Braatvedt G, Hurt DE, Katsantoni E, Sertedaki A, Chrousos GP and Charmandari E: A novel point mutation of the human glucocorticoid receptor gene causes primary generalized glucocorticoid resistance through impaired interaction with the LXXLL motif of the p160 coactivators: Dissociation of the transactivating and transreppressive activities. J Clin Endocrinol Metab. 99:E902–E907. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Jääskeläinen J, Mongan NP, Harland S and Hughes IA: Five novel androgen receptor gene mutations associated with complete androgen insensitivity syndrome. Hum Mutat. 27(291)2006.PubMed/NCBI View Article : Google Scholar | |
|
Vitellius G, Fagart J, Delemer B, Amazit L, Ramos N, Bouligand J, Le Billan F, Castinetti F, Guiochon-Mantel A, Trabado S, et al: Three Novel Heterozygous Point Mutations of NR3C1 Causing Glucocorticoid Resistance. Hum Mutat. 37:794–803. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Harrod A, Fulton J, Nguyen VTM, et al: Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer. Oncogene. 36:2286–2296. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Achermann JC, Schwabe J, Fairall L and Chatterjee K: Genetic disorders of nuclear receptors. J Clin Invest. 127:1181–1192. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Ai N, Krasowski MD, Welsh WJ and Ekins S: Understanding nuclear receptors using computational methods. Drug Discov Today. 14:486–494. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Matias PM, Carrondo MA, Coelho R, Thomaz M, Zhao XY, Wegg A, Crusius K, Egner U and Donner P: Structural basis for the glucocorticoid response in a mutant human androgen receptor (AR(ccr)) derived from an androgen-independent prostate cancer. J Med Chem. 45:1439–1446. 2002.PubMed/NCBI View Article : Google Scholar | |
|
Puyang X, Furman C, Zheng GZ, Wu ZJ, Banka D, Aithal K, Agoulnik S, Bolduc DM, Buonamici S, Caleb B, et al: Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ERαWT and ERαMUT Breast Cancer. Cancer Discov. 8:1176–1193. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Nettles KW, Bruning JB, Gil G, O'Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, et al: Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 8:563–568. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Siltberg-Liberles J, Grahnen JA and Liberles DA: The evolution of protein structures and structural ensembles under functional constraint. Genes (Basel). 2:748–762. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D and Gronemeyer H: A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 3:87–94. 1996.PubMed/NCBI View Article : Google Scholar | |
|
Kandil S, Biondaro S, Vlachakis D, Cummins AC, Coluccia A, Berry C, Leyssen P, Neyts J and Brancale A: Discovery of a novel HCV helicase inhibitor by a de novo drug design approach. Bioorg Med Chem Lett. 19:2935–2937. 2009.PubMed/NCBI View Article : Google Scholar |