microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer
- Authors:
- Published online on: December 28, 2011 https://doi.org/10.3892/etm.2011.436
- Pages: 560-566
Abstract
Introduction
microRNAs (miRNAs) are endogenous small single-strand non-coding RNAs of approximately 22 nucleotides (1) that have emerged as a prominent class of gene regulators. miRNAs may have causal roles in numerous normal cellular and tumor processes, such as development, differentiation, proliferation and apoptosis (2,3). Moreover, a growing number of miRNAs have been classified as oncogenes or tumor-suppressor genes (4,5). Over the past decade, an increasing number of studies have shown dysregulation of miRNA expression in numerous tumor types including esophageal (6), lung (7), liver (8), pancreatic (9), bladder (10), ovarian (11) and gastric cancer (12).
In previous studies, the evidence has revealed that several miRNA expression levels were aberrant in colorectal cancer (CRC). Cummins et al (13) indicated that 53 miRNAs were expressed at significantly different levels between CRC tissues and normal colonic epithelium by microarray and qRT-PCR methods. Moreover, Chen et al (14) indicated that miR-148a and -152 were downregulated, as shown by real-time PCR assay, in a large number of cases with CRC and were significantly related to tumor size and depth of invasion. Our previous study showed similar results, in that miR-203 was significantly downregulated in CRC by real-time PCR assay. Also, the low expression of miR-203 was correlated with increased tumor size and advanced depth of invasion (15). Furthermore, Georges et al (16) reported that miR-192 and -215 were downregulated by microarray and qRT-PCR methods in human CRC cell lines, and these miRNAs, as tumor suppressors, led to cell cycle arrest.
In this study, we detected the expression levels of miR-192, -194 and -215 in a large number of CRC tissues, relative to their non-tumor counterparts, by real-time PCR assay, as well as in three CRC cell lines. Further investigation revealed that aberrant expression of these miRNAs was, notably, correlated with clinicopathological characteristics in CRC. Moreover, we studied the association between miR-194 and cell proliferation in vitro by MTT assay. Our findings will help to elucidate the functions of miRNAs and their role in carcinogenesis.
Material and methods
Tissues samples
A total of 107 pairs of CRC tissues and non-tumor adjacent tissues (NATs; as the controls) were obtained from patients that underwent radical resection between 2007 and 2010 at the First Hospital of China Medical University (Shenyang, China). The non-tumor counterparts were obtained from a section of the resected specimen at the farthest distance from the tumor. The samples were snap-frozen in liquid nitrogen immediately following surgery and were stored at −80°C until use. No previous local or systemic treatment had been conducted on these patients prior to surgery.
CRC was subsequently diagnosed based on histopathological evaluation. One section of each sample was stained with hematoxylin-eosin (H&E). The histological grade of cancer was classified using the TNM staging system of the American Joint Committee on Cancer (AJCC; 2010) and the International Union Against Cancer (UICC), according to the standard of the World Health Organization (WHO). Informed consent was obtained from all patients. The study was approved by the Research Ethics Committee of China Medical University (Shenyang, China).
Cell lines
Human CRC cell lines (HT-29, HCT-116 and SW-620) were obtained from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). HT-29 and HCT-116 were cultured in McCoy's 5A medium (Invitrogen, Carlsbad, CA, USA); SW-620 was cultured in Leibovitz's L-15 medium (Invitrogen). All of the cell lines were cultured at 37°C in a humidified atmosphere of 5% CO2. Media were supplemented with 10% fetal bovine serum (FBS).
RNA isolation and reverse transcription reaction
Total RNA was isolated from the specimens using a mirVana miRNA Isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. A UV spectrophotometry NanoPhotometer UV/Vis spectrophotometer (Implen, Schatzbogen, München, Germany) was used to determine the concentration and purity of RNA. A poly(A) tail was added to RNA in a 37°C water bath for 30 min by Escherichia coli poly(A) polymerase (E-PAP) using a Poly(A) Tailing kit, according to the manufacturer's instructions (Ambion) (17). Following purification by phenol-chloroform and ethanol, RNAs were dissolved in diethyl pyrocarbonate (DEPC)-treated water. The first-strand cDNA was synthesized with the SuperScript® III First-Strand Synthesis System using a reverse transcription-polymerase chain reaction kit (Invitrogen). To generate cDNA of miRNA, a 10 μl reverse transcription reaction mixture containing 1 μg of the RNA sample, 1 μl RT-primer (Table I), 1 μl 10 mM deoxyribonucleotide triphosphate (dNTP) mix and DEPC-treated water at 65°C was incubated for 5 min. Then, a 10 μl mixture containing 2 μl 10X RT buffer, 4 μl 25 mM MgCl2, 2 μl 0.1 M DTT, 1 μl RNaseOUT (40 U/μl) and 1 μl SuperScript III RT (200 U/μl) was added. The total reaction mixture was incubated in a 96-well plate of a GeneAmp PCR 9700 Thermocycler (Applied Biosystems, Hayward, CA, USA) for 50 min at 50°C, 5 min at 85°C, and 20 min at 37°C after adding 1 μl RNase H to the mixture, and held at 4°C.
Table I.RT-PCR primers for amplification of miR-192, -194 and -215 expression and the sequences of miR-194 mimics and NC. |
Real-time PCR
According to the manufacturer's instructions, real-time PCR was performed using the SYBR Premix Ex Taq™ II kit (Takara Bio, Kyoto, Japan) with a Rotor-gene 6000 system (Qiagen, Valencia, CA, USA) (17). The 25 μl mixture of PCR consisted of 12.5 μl SYBR Green supermix, 8.5 μl RNase-free water, 1 μl forward primers, 1 μl reverse primers and 2 μl reverse transcribed product. Threshold cycle data were determined by setting a default threshold. The reactive condition was 45 amplification cycles of 95°C for 5 sec, 58°C for 20 sec and 72°C for 30 sec in a 36-well optical plate using a Rotor-gene 6000 system. The U6 RNA was selected as an endogenous reference to calculate the relative expression levels of miR-192, -194 and -215 in cancerous samples compared to non-tumor counterparts using the 2−ΔΔCt method (18). All samples were performed in triplicate and repeated three times. The products of real-time PCR were confirmed by TA cloning and a sequencing assay. The primers for miR-192, -194 and -215 and the endogenous control U6 are shown in Table I.
Cell transfection and MTT assay
miR-194 mimics were composed of an RNA duplex (Table I) designed as described previously (19). Non-specific sequences were non-homologous to any human genome sequences as a negative control RNA duplex (named as NC, Table I). Corresponding 2-O-methyl analogues were used to substitute for all pyrimidine nucleotides in the miR-194 mimics or NC to improve RNA stability for an MTT assay in vitro. miR-194 mimics (50 nM) and NC were transiently transfected in cultured SGC-7901 cells at 30-50% confluence using Lipofectamine 2000 (Invitrogen). All the RNA oligoribonucleotides were chemically synthesized by GenePharma (Shanghai, China).
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed to measure the capacity for cellular proliferation. A total of 24 h following transfection, cells (approximately 0.4×104) were seeded into 96-well microtiter plates for 24, 48, 72 and 96 h. Then, the cells were incubated with 20 μl of MTT (5 mg/ml) for 4 h at 37°C and 150 μl of dimethyl sulfoxide (DMSO) was added to solubilize the crystals for 20 min at room temperature. A spectrophotometer (Multiskan MK3; Thermo, Waltham, MA, USA) was used to measure the optical density (OD) at a wavelength of 490 nm. All experiments were performed three times to calculate the average results. The growth inhibition rate was calculated as follows: (AC - AT)/AC x 100% (AC = absorbance value of the NC; AT = absorbance value of the experimental group) (20).
Statistical analysis
For real-time PCR, the threshold cycle of fluorescence (Ct) for each sample was determined to evaluate the association between CRC tissues and matched NATs by the 2−ΔΔCt method. ΔΔCt indicates the difference in the ΔCt value between cancer tissue and the corresponding control (ΔΔCt = ΔCt cancer − ΔCt control) and ΔCt is the difference of the Ct value between the target and U6 (ΔCt = Ct target − Ct U6). Finally, the 2−ΔΔCt value (fold value) was calculated and distinguished as 1-fold, and a fold value of less than 1-fold was defined as low expression (15,21). Differences in miRNA expression were measured by comparing the values of ΔCt cancer and ΔCt control, and statistical differences in miRNA expression levels were determined using a paired t-test in cancer tissues and cancer cell lines relative to non-tumor counterparts, as well as comparing the effect of miR-194 on cell proliferation in HCT-116 cells by MTT assay. Moreover, the association between miRNA expression levels and clinicopathological parameters was analyzed by a non-parametric test (Mann-Whitney U test between two groups and Kruskal-Wallis H test for three or more groups). P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using the Statistical Program for Social Sciences (SPSS) software 16.0 (SPSS Incorporated, Chicago, IL, USA).
Results
Expression of miR-192, -194 and -215 in CRC
Among 107 patients with CRC, expression levels of miR-192, -194 and -215 were detected using real-time PCR in cancer tissues compared to matched non-tumor counterparts and the values of ΔCt (means ± SD) were 4.632±2.090, 4.680±2.170 and 6.483±2.242 in cancer tissues, and 2.207±2.704, 2.780±2.922 and 4.032±2.768 in non-tumor counterparts, respectively. Moreover, miR-192, -194 and -215 were significantly downregulated in CRC tissues with the median 0.13-fold, 0.23-fold, and 0.15-fold relative to the control group, respectively (all p<0.001, paired t-test; Fig. 1). Furthermore, in 87 of 107 (81.30%), 85 of 107 (79.44%) and 86 of 107 (80.37%) cases, miR-192, -194 and -215 expression levels revealed a >50% reduction between the two types of tissues. In the cell lines, we also found a significantly lower expression of miR-192, -194 and -215 in HT-29 cells (p=0.003, p=0.016, and p=0.002, respectively; paired t-test), HCT-116 cells (p=0.003, p=0.014 and p=0.001, respectively) and SW-620 cells (p=0.002, p=0.010 and p=0.002, respectively) compared to normal colorectal tissues (Fig. 2).
Association between expression of miRNAs and clinicopathological characteristics in CRC
In our study, there was an association between miRNA expression levels and clinicopathological characteristics in CRC. Lower expression levels of miR-192, -194 and -215 in patients with CRC tended to be associated with increased tumor sizes as shown by non-parametric tests (p=0.027, p=0.018 and p=0.027, respectively; Mann-Whitney U test, Table II). There was no significant difference between low expression of the miRNAs and other clinicopathological characteristics such as gender, age, histological grade, pT stage, pN stage, clinical stage, lymph node metastasis rate and lymphatic vessel invasion.
Table II.Association between miR-192, -194 and -215 expression and clinicopathological features in patients with colorectal cancer. |
Marked correlations among miR-192, -194 and -215
There were marked correlations between miR-192 and -194, miR-192 and -215, and miR-194 and -215 in CRC tissues, which were evaluated by Pearson's regression analysis (all p<0.001; Fig. 3). The correlation coefficients were 0.950, 0.895, and 0.856, respectively.
Effects of miR-194 on cell proliferation
The effect of miR-194 on cell proliferation was assessed in HCT-116 cells. The transfection efficiency was detected by the real-time PCR method 48 h following transfection. Moreover, we found that the cells that were transfected with miR-194 mimics in HCT-116 cells had marked growth inhibition at the point of 96 h post-transfection compared to the matched NC and SCG-7901 cells, as shown by the MTT assay (Fig. 4). Furthermore, the inhibition rate was 19.20% at the point of 96 h post-transfection in HCT-116 cells.
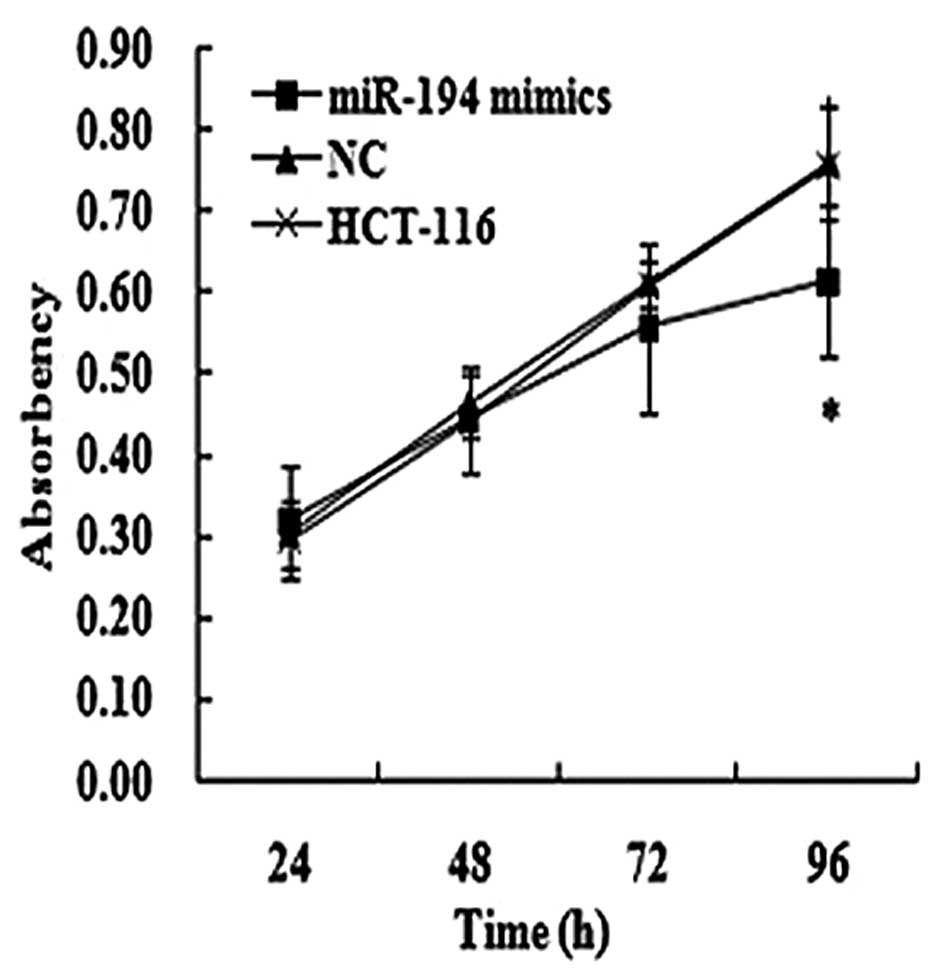 | Figure 4.miR-194 significantly inhibited cell proliferation in HCT-116 cells by MTT assay (*P<0.05). |
Discussion
In recent years, studies have shown that miRNA alterations may function as a novel class of oncogenes and tumor suppressors, which could be used for the diagnosis and treatment of cancer (22). Mathé et al (6) reported that miR-192 and -194 were upregulated in 107 patients with esophageal cancer compared to non-tumor counterparts by microarray assay. Moreover, Jin et al (23) indicated that miR-192 and -215 in gastric cancer tissues and cell lines were upregulated by microarray and qRT-PCR methods. Furthermore, more evidence revealed the upregulated miR-192 in lung cancer, overexpression of miR-194 in highly metastatic pancreatic ductal adenocarcinoma, and overexpression of miR-215 in hepatocellular carcinoma by microarray and real-time PCR methods (7–9). However, miR-194 in mouse hepatocellular carcinoma and cancer cell lines was downregulated by the real-time PCR method (24). Therefore, the altered miRNA expression levels may cause tissue-specific differences. Just as Baffa et al suggested, the various miRNA expression levels, which were observed in different organs of origin, were markedly tissue-specific (25). In the future, the correlation between miRNAs and cancer could become a focus of cancer studies.
miRNAs can be quantified by microarray assay, bead-based flow cytometric assay and real-time PCR assay. The main advantage of real-time PCR is that it is more quantitative and sensitive than other assays. Therefore, we performed real-time PCR on a large number of cases to assess the expression levels of miR-192, -194 and -215 in CRC. The significantly lower expression of these miRNAs was found in 107 cancer tissues compared to non-tumor counterparts. Moreover, research on the miRNA expression levels in CRC cell lines also provided similar significant results. In previous studies, similar results indicated that miR-192, -194 and -215 were downregulated in CRC cell lines and rat colon tissues (16,26,27). Furthermore, our studies revealed that the increased tumor size in CRC was closely correlated with the low expression of miR-192, -194 and -215. There was also an inverse correlation between tumor size and miRNA expression levels, with lower expression levels being associated with increased tumor size. Although the increased tumor size in CRC was not part of the staging system, previous studies have suggested that tumor size is also an important prognostic factor in CRC (28). Thus, miR-192, -194 and -215 might be important biological markers in the carcinogenesis of CRC and the low expression of these miRNAs may contribute to the proliferation of CRC.
In the present study, we found a marked correlation between miR-192 and -194, miR-192 and -215, and miR-194 and -215 in CRC tissues. As shown on the miRBase and HGNC website, miR-192 and -215 have the same ‘seed region’; miR-192 and -194-2 are on the same chromosome at the 11q13.1 and miR-194-1 and -215 are on 1q41. Moreover, the mature sequence of miR-194-1 and -194-2 is miR-194. We therefore suggest that miRNAs may contribute to the carcinogenesis of CRC in synergism. Confirmation of this theory requires further investigation.
In a previous study, Boni et al indicated that miR-192 and -215 in DLD-1 CRC cell lines induced the accumulation of p53 and suppressed cell proliferation in a partially, but not completely, p53-dependent pathway (29). Furthermore, Song et al also found similar results in HCT-116 CRC cell lines transfected with miR-215. Their results suggest that the reduced proliferation rate is due to a decreased S phase and increased G2 checkpoint control (30). More evidence was observed that miR-192 and -215 may function as tumor suppressors capable of inhibiting cell proliferation, suppressing carcinogenesis through p21 accumulation, and causing cell cycle arrest in CRC cell lines (16,26,31). In the present study, cell proliferation assays were performed in HTC-116 cells to investigate the potential impact of miR-194 on cell growth. Our results showed that miR-194 mimics transfection in HCT-116 cells was significantly lower than the NC group and blank group by MTT assay. Thus, we suggest that the overexpression of miR-194 may be a potential biological marker for the inhibition of cell proliferation in CRC. Future studies on the functions of these miRNAs are required to further the investigation of CRC.
There are a number of factors that may reduce miRNA expression, including transcriptional factors, mutations, deletions and methylation. Recent studies have shown that hypermethylation of the miR-194-2 and -192 cluster promoter in multiple myeloma (MM) cell lines suggests that epigenetic downregulation of these miRNAs, which leads to an increase in murine double minute 2 (MDM2) mRNA and protein expression, decreases the ability of p53 to downmodulate MDM2 expression (32). Moreover, Hino et al (33) indicated that miR-194 was transcriptionally upregulated in a gastrointestinal tract enriched nuclear receptor by the hepatic nuclear factor 1α (HNF1-α) in intestinal epithelial cells. Therefore, considering these reasons, we speculate that hypermethylation and transcription factors may be mechanisms for the downregulation of miR-192, -194 and -215 in CRC.
In a large number of CRC tissues, miR-192, -194 and -215 were significantly downregulated relative to their non-tumor counterparts, as well as in the three CRC cell lines. Also, a significant association between these miRNA expression levels and increased tumor size was found in CRC. Moreover, there was a marked correlation among these miRNAs in CRC tissues. An in vitro cell proliferation assay revealed that the miR-194 mimics transfection in HCT-116 cells was significantly lower than controls. The present study indicates a basis for further studies on target genes and identification of more functions of these miRNAs in CRC. In future studies, the associations between these miRNAs and the prognosis of patients with CRC need to be confirmed using large-scale and long-term follow-up studies.
Acknowledgements
This study was supported by grants from the National Science Foundation of China (No. 30972879 and No. 81000943), Specialized Research Fund for the Doctoral Program of Higher Education (No. 200801590006) and Natural Science Foundation of Liaoning Province (No. 20092129).












