Comprehensive analysis of histone modification‑associated genes on differential gene expression and prognosis in gastric cancer
- Authors:
- Published online on: July 24, 2019 https://doi.org/10.3892/etm.2019.7808
- Pages: 2219-2230
Abstract
Introduction
Gastric carcinoma (GC) is the second most common human cancer type and a leading cause of cancer-associated mortality worldwide. Although the incidence has significantly declined due to recent advances in diagnostics and therapeutics, the survival rate remains poor. Multistep processes, including genetics, epigenetics and environmental factors, have pivotal roles in tumorigenesis and progression (1). The identification of novel biomarkers in the above processes for clinical applications is urgently required.
The histone core proteins (two of each H2A, H2B, H3 and H4) together with 146 bp DNA are wrapped around each other and form a nucleosome, which constitutes the basic units of chromatin (2). In dynamic and reversible processes, as the chromosomes are condensed or loosened, the N-terminus of the histone proteins may be altered by multiple covalent modifications, including acetylation, methylation, phosphorylation and ubiquitination, at the post-transcriptional level to regulate gene expression. Numerous studies on covalent modifications have focused on exploring the roles of acetylation and methylation (3,4). Dysregulations of these epigenetic modifications and associated gene expression causes may drive carcinogenesis. Downregulation of lysine acetyltransferase 5 (KAT5) (5) and upregulation of enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) (6), protein arginine methyltransferase 1 (PRMT1) (7) and the lysine demethylase 1A (KDM1A) (8) have been reported to be significantly associated with poor clinicopathological features and survival of patients with GC and other cancer types. As one of the epigenetic mechanisms, histone modifications participate in transcriptional regulation, DNA repair and condensation (9). Histone deacetylase 4 has been reported to facilitate GC progression by inhibiting p21 (10). P300 acetylates transcription factor (TF) STAT3 in histone H3 on lysine 56 to regulate gene expression (11). Furthermore, EZH2 recruits DNA methyltransferase to the promoter region of PcG target genes to downregulate PcG targets (12). In addition, lysine demethylase 6B interacts with NF-κB via demethylation of histone H3 trimethylated at lysine 27 (H3K27me3) at downstream gene promoters participating in wound healing (13). In the DNA damage repair process, KAT8 was also reported to be required (14). Alterations in histone modification levels and the expression of numerous genes encoding histone modification-associated enzymes have also been reported in various cancer types as epigenetic changes. For instance, SUV420H2 (KMT5C)-mediated histone H4 trimethylation on lysine 20 is important for epidermal homeostasis. SET domain bifurcated histone lysine methyltransferase 1, which performs H3K9 trimethylation (H3K9me3), is established as an oncogene in melanoma (15). Upregulation of JMJD3 in metastatic prostate cancer indicates its potential oncogenic role (16). Similarly, KDM1A and JMJD1C are upregulated in certain GC cells and tissues (17). Therefore, histone modification-associated changes may serve as biomarkers for early diagnosis, therapeutic targets and prognosis prediction of GC.
Although a large number of studies have indicated that histone modifications and abnormal expression of associated genes have important roles in oncogenesis, only few studies have provided comprehensive analyses of the expression and prognostic value of associated genes in carcinomas, particularly in GC. The present study analyzed Oncomine, The Cancer Genome Atlas (TCGA) and Gene Ontology (GEO) datasets to perform comprehensive analyses on histone modifications and associated gene expression profiles, as well as their prognostic role in GC.
Materials and methods
Oncomine database analysis
The Oncomine database (http://www.oncomine.org) incorporates 715 datasets that include 35 cancer types and contain microarray data of 86,733 samples, supporting various methods of online statistical analysis (18). The differential expression of histone modification and associated genes (HMGs) was compared by using the Student's t-test to generate a P-value. As cut-off values, the |logFC| was defined as >1 and the P-value was set at 0.05, whereas the data type was restricted to mRNA.
TCGA and GEO datasets
The UALCAN database (http://ualcan.path.uab.edu/index.html) was used to screen the HMGs (19). The mRNA expression datasets from the TCGA stomach adenocarcinoma (STAD) dataset, which contained 375 GC samples and 32 normal samples, were downloaded for further analysis (http://cancergenome.nih.gov/). The clinical data of 443 patients with GC, including overall survival (OS), survival state, age, sex, location, American Joint Committee on Cancer (AJCC) Tumor-Nodes-Metastasis (TNM) stage and pathological T/N/M stage were also obtained/estimated. The integrated above data were used to assess the association between mRNA expression levels and prognosis. The mRNA microarray expression profile dataset GSE79973 was obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) and contained 10 GC samples and 10 paired normal samples.
Kaplan-Meier plotter database analysis
Kaplan-Meier plotter is capable of assessing the effect of 54,675 genes on survival using 10,461 cancer samples. These include the data of 5,143 breast, 1,816 ovarian, 2,437 lung and 1,065 GC patients. The association between the expression of certain genes and survival of GC patients was analyzed using Kaplan-Meier plotter (http://kmplot.com/analysis/) (20). The hazard ratios (HRs) with 95% confidence intervals (CI) and log-rank P-values were also computed.
Statistical analysis
The association between histone modifications and the expression of relevant genes as well as clinicopathological features was evaluated by the χ2 test. An unpaired t-test was used to compare the differentially expressed genes (DEGs) between the tumor and non-tumor group. Spearman's test was used for correlation analysis. The Kaplan-Meier method was used to determine the patients' survival and differences between groups were assessed using a log-rank test. In TCGA dataset, statistically significant variables in the univariate analysis were included into the multivariate analysis in the Cox proportional hazards model and results were expressed as the HR with 95% CI. Data analysis was performed using SPSS software v.23.0 (IBM Corp.). The mRNA data downloaded from TCGA and GEO were analyzed using the edgeR package in R (v.3.5.1) to identify DEGs between GC and non-tumor tissues. The median value of mRNA expression was applied as a cut-off to stratify samples into high- or low-expression groups. The STRING v.10.5 online tool (https://string-db.org/) was used to analyze the interactions among differential proteins. All of the P-values reported were two-sided and P<0.05 was considered to indicate statistical significance.
Results
Screening and identification of DEGs in the HMGs from Oncomine, GEO and TCGA datasets
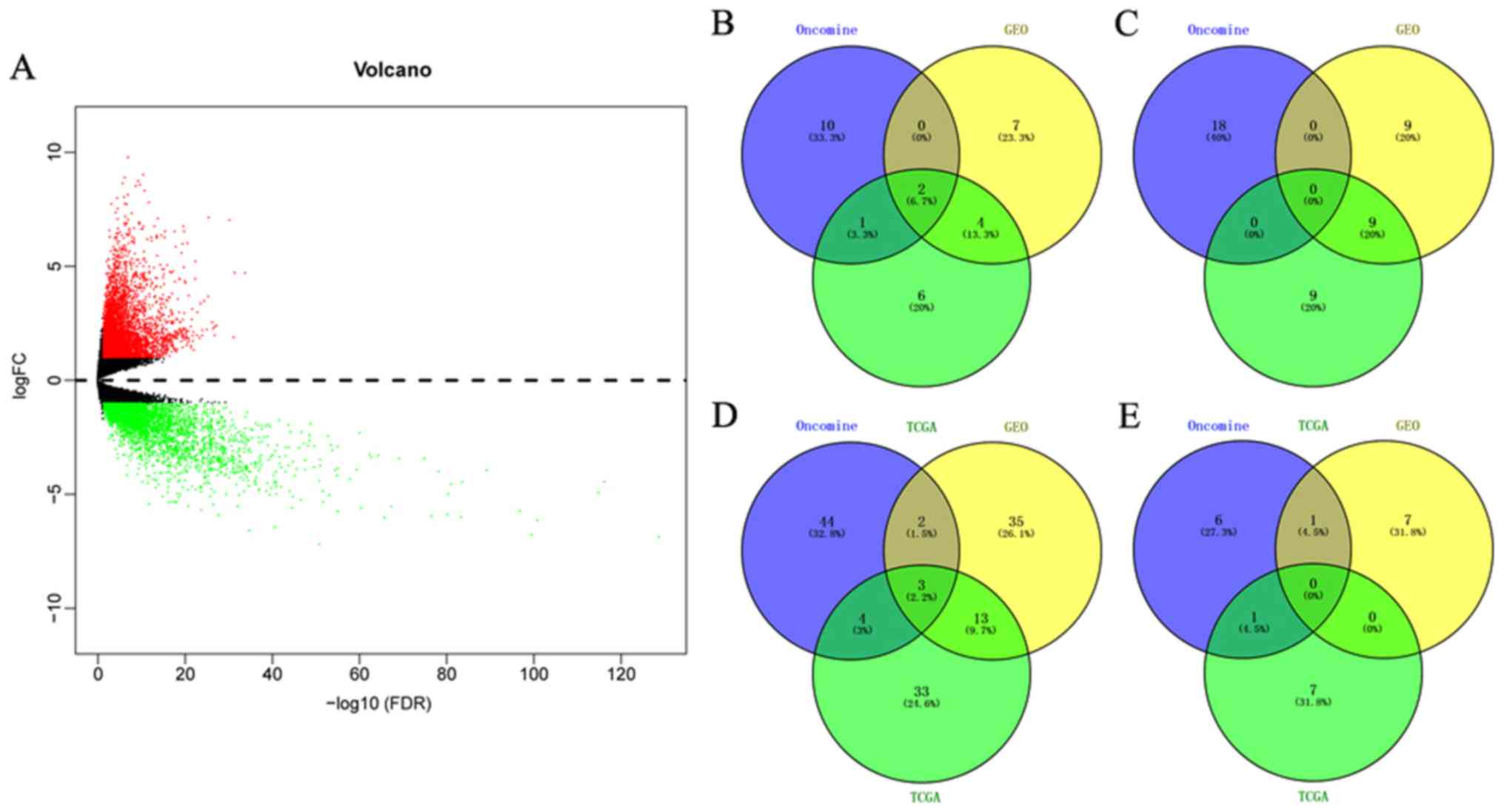
The microarray expression profile dataset GSE79973 and mRNA expression data were respectively downloaded from the GEO and TCGA databases. DEGs were screened out using the cutoffs of P<0.05 and |logFC|>1 for the comparison between tumor and non-tumor samples. A total of 1,311 upregulated and 384 downregulated genes screened from GSE79973 (Fig. S1) and 8,159 upregulated and 3,758 downregulated genes were screened from TCGA (Fig. 1A). The HMGs were then retrieved and determined using the UALCAN website. A total of 30 genes involved the acetyl modification of histones (12 acetyltransferases and associated genes, 18 deacetylases and associated genes) and 61 genes participating in methyl modification (53 methyltransferases and associated genes, 8 demethylases and associated genes) were identified. Detailed information on the location and function of the HMGs is provided in supplementary Table SI. The differences in expression from HMGs between various carcinomas and paracancerous tissues were also indicated in the Oncomine analysis (Fig. S2).
To screen the HMGs that were differentially expressed in GC vs. non-cancer tissues among all Oncomine, GEO and TCGA datasets, Venn diagram analysis was used to obtain the intersection of the three datasets. As presented in Fig. 1B-E, the five genes KAT2A, nuclear receptor coactivator 1 (NCOA1), SMYD family member 5 (SMYD5), PRMT1 and PR/SET domain 16 (PRDM16) were identified. Detailed information on the location and function of these HMGs is provided in Table I.
Table I.Basic characteristics and function of five histone modification enzymes and associated genes. |
Differential expression of KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 in GC vs. normal tissues
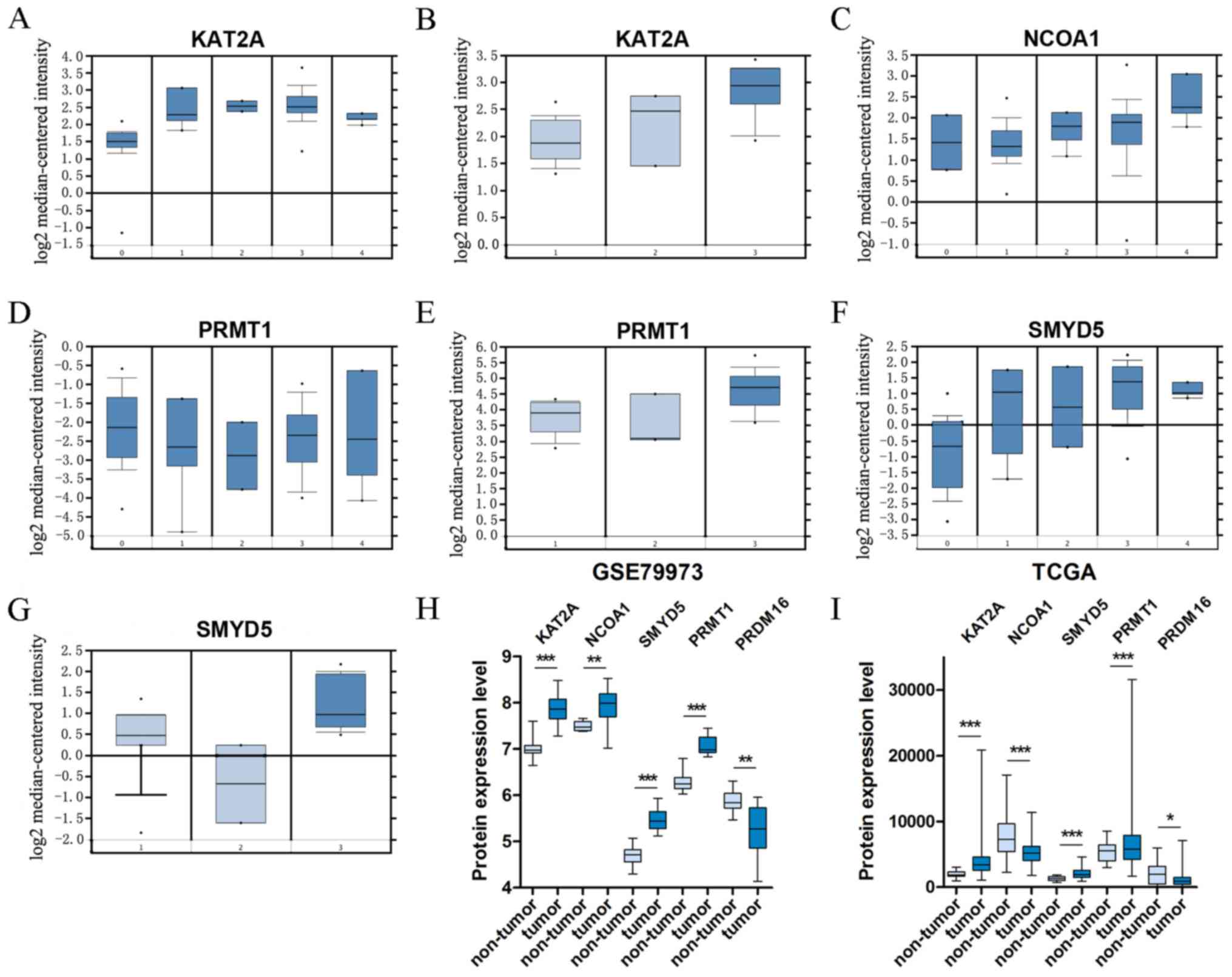
KAT2A, SMYD5 and PRMT1 were upregulated in GC vs. normal tissues in all three datasets (Oncomine, GSE79973 and TCGA). In the GSE79973 and TCGA STAD datasets, PRDM16 was observed to be downregulated. Furthermore, in the GSE79973 dataset and the dataset DErrico Gastric (Oncomine dataset), NCOA1 was upregulated, and these results were contrary to those obtained with the TCGA STAD dataset (Table II). A 29% decline of NCOA1 mRNA expression was observed in TCGA STAD dataset. In the Wang Gastric and Cui Gastric datasets (Oncomine dataset), KAT2A was 1.756-fold (P=1.15×10−4) and 1.461-fold (P=1.37×10−5) increased, respectively, in GC vs. normal tissues. Furthermore, the FC of upregulated KAT2A in the GSE79973 dataset was 1.809 (P=1.89×10−5). In the Wang Gastric dataset from Oncomine, SMYD5 and PRMT1, which in turn was also determined to be 1.470-fold (P=1.00×10−3) increased in the Cui Gastric dataset, were 2.154-fold (P=6.18×10−4) and 1.884-fold (P=8.01×10−4) elevated, respectively. Similar to the results of the Oncomine analysis, screening of the GSE79973 dataset indicated that NCOA1, SMYD5 and PRMT1 were 1.347-fold (P=8.95×10−3), 1.708-fold (P=7.39×10−6) and 1.724-fold (P=2.00×10−6) increased. Furthermore, comparisons based on different pathological classifications were performed to determine differential expression of KAT2A, NCOA1, SMYD5 and PRMT1, and in the datasets DErrico Gastric and Cho Gastric in the Oncomine analysis, the FC of KAT2A, NCOA1, SMYD5 and PRMT1 in gastric intestinal-type adenocarcinoma was higher than in gastric mixed and diffuse gastric adenocarcinoma compared to normal tissues (respective FCs and P-values: 2.132, 1.61×10−11; 1.562, 7.00×10−3; 4.106, 1.08×10−10; 2.776, 2.62×10−11). The box plots in Fig. 2A-I display the significant alterations in expression among the five genes in subgroup comparisons.
Table II.Differential expression of five histone modification enzymes and associated genes between different types of gastric cancer and normal tissues. |
Association of KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 expression with clinicopathological features
To explore the potential clinical significance of the five DEGs in patients with GC, the clinical data of 443 patients were downloaded from TCGA and only 334 patients with complete clinical and gene data were retained after elimination of patients with missing data by processing in R. The cases were divided into a low-expression group and a high-expression group according to the median mRNA expression value of the five genes in tumor or non-tumor tissues. The associations between the expression of the five genes and clinicopathological characteristics are presented in Table III. Factors including age, sex, tumor size, AJCC stage and T/N/M stage were evaluated. Unfortunately, no significant associations were observed between the five aberrantly expressed genes and the above-mentioned clinical features.
Table III.Association between five histone modification enzymes and associated genes and clinicopathological parameters of gastric cancer patients. |
Prognostic value of KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 in GC
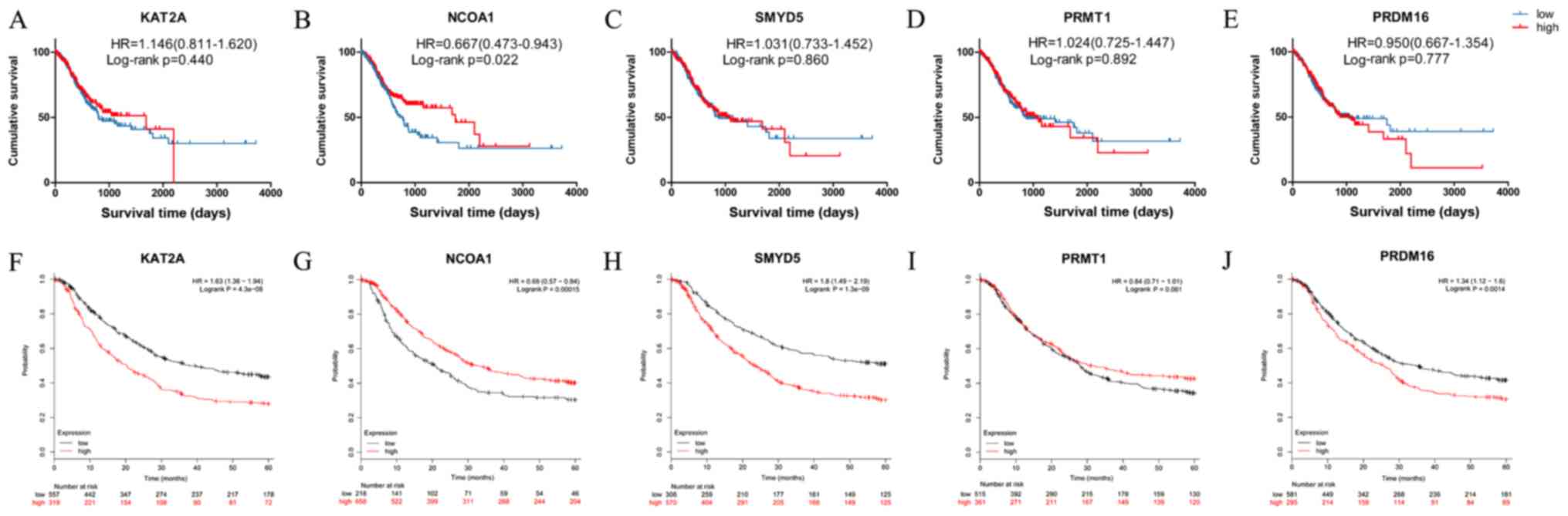
The prognostic value of the expression status of KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 was first examined in the TCGA dataset (Fig. 3A-E). Low mRNA expression of NCOA1 mRNA was observed to be linked to significantly better OS in GC (HR=0.667, 95% CI=0.473–0.943, P=0.022; Fig. 3B). However, the mRNA expression status of KAT2A, SMYD5, PRMT1 and PRDM16 mRNA was not associated with OS in GC (HR=1.146, 95% CI=0.811–1.620, P=0.440; HR=1.031, 95% CI=0.733–1.452, P=0.860; HR=1.024, 95% CI=0.725–1.447, P=0.892; HR=0.950, 95% CI=0.667–1.354, P=0.777; Fig. 3A and C-E, respectively). Kaplan-Meier plotter analysis was also used to verify the associations between the mRNA expression of five genes and the clinical survival outcome (Fig. 3F-J). In order to reduce the bias caused by the conversion of time (day or mouth), the time units were used as presented in the respective datasets for the Kaplan-Meier survival curves. As in the TCGA dataset, low NCOA1 expression in GC tissues was associated with better OS (HR=0.690, 95% CI=0.570–0.840, P<0.001), and the survival curve for the group with high PRMT1 mRNA expression in GC tissues did not exhibit any significant difference from that of the low expression group (HR=0.840, 95% CI=0.710–1.010, P=0.061; Fig. 3I). Contrary to the results obtained with the TCGA dataset, patients with high expression of KAT2A, SMYT5 and PRDM16 in their GC tissue exhibited significantly poorer OS than those with corresponding low expression according to Kaplan-Meier plotter analysis (Fig. 3F, H and J, respectively). The Cox proportional hazards model indicated that NCOA1 expression, age, N stage, M stage were independent predictors for survival in GC (HR=1.523, 95% CI=1.072–2.163, P=0.019; HR=1.939, 95% CI=1.291–2.914, P=0.001; HR=1.233, 95% CI=1.049–1.450, HR=2.419, 95% CI=1.423–4.115, respectively; Table IV).
Bromodomain protein ATPase family AAA domain- containing protein 2 (ATAD2) is associated with the HMGs
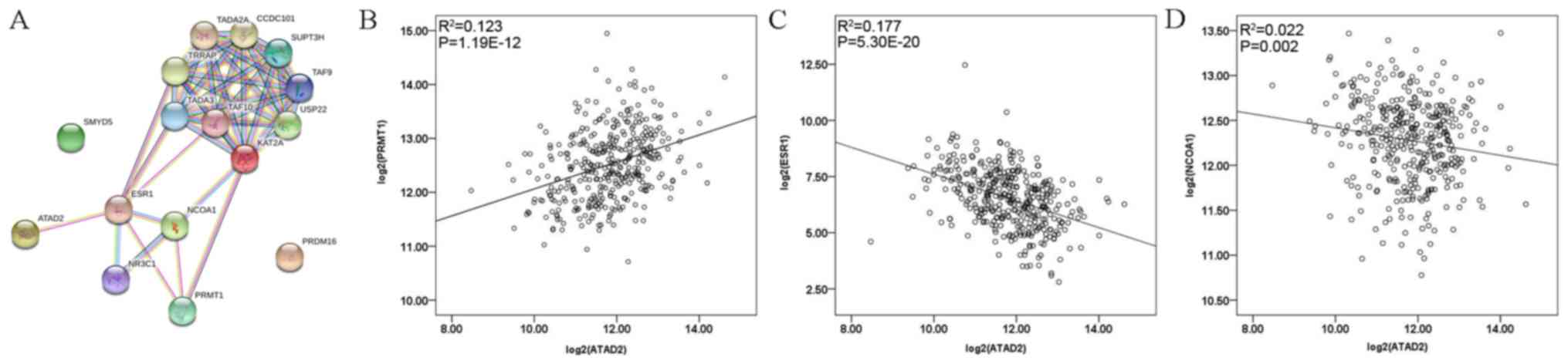
ATAD2, a member of the AAA + ATPase family of proteins, contains a bromodomain and its functions are linked to genome regulation and histone modification. A previous study by our group on hepatocellular carcinoma suggested that ATAD2 was overexpressed in tumor tissue and associated with poor survival (21). To explore the association between ATAD2 and the five genes screened out from datasets in the present study, the STRING v.10.5 online tool (https://string-db.org/) was used to display the interactions among them. As presented in Fig. 4A, protein-protein interaction network highlighted the association between ATAD2 and estrogen receptor 1 (ESR1), PRMT1, NCOA1 and KAT2A. Associations between clinicopathological features and ATAD2 or ESR1 mRNA expression were also assessed (Table SII) and no statistical significances were observed. Spearman's correlation analysis revealed that ATAD2 expression was positively correlated with PRMT1 (R2=0.123, Spearman rho=0.356, P=1.19×10−12; Fig. 4B) and negatively correlated with ESR1 (R2=0.177, Spearman rho=−0.449, P=5.30×10−20; Fig. 4C) and NCOA1 (R2=0.022, Spearman rho=−0.160, P=0.002; Fig. 4D).
Discussion
The maintenance of stable and ordered chromatin during dynamic packaging is vital to normal cellular homeostasis. Warped histones and DNA are subject to covalent post-translational modifications in order to influence the number of chromatin-associated cellular events, including transcription, replication recombination and DNA repair (22). Dysregulation of the epigenetic mechanisms that govern transcriptional regulation resembles steps in the oncogenic process, causing inappropriate activation of oncogenes or the inhibition of tumor suppressors and leading to carcinogenesis. Accumulating evidence has demonstrated that epigenetic alterations caused by histone modifications also have important roles in gastric carcinogenesis, particularly the well-studied acetylation and methylation modifications (23). The histone modifications-associated enzymes and encoding genes are among the most frequent abnormal targets in aberrant histone modifications. Genes that encode histone modification enzymes, including the CREB binding protein (CBP), p300, KAT5, KDM1A and JMJD1C, have been reported to be aberrantly expressed in GC and are significantly correlated with poor survival (5,6,17,24–26). Possibly due to lack of data at the genomic level, comprehensive molecular characterization of histone modification regulations in GC has rarely been performed. To the best of our knowledge, the present study was the first to elucidate histone modifications, associated gene expression profiles and prognostic roles of key genes by using Bioinformatics analysis of datasets to explore the implication of deregulated histone modification in the initiation and development of GC and the underlying mechanisms.
The present results suggested that KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 were differentially expressed in GC vs. non-cancer tissues. Among them, KAT2A, SMYD5 and PRMT1 were highly upregulated in GC compared with normal tissues. The expression of PRDM16 was observed to be downregulated in GC. However, the opposite result was identified when detecting NCOA1 expression. Analysis of the GEO and Oncomine datasets revealed that NCOA1 was upregulated; these results were contrary to those of the TCGA STAD dataset. As a member of the p160 SRC family, which also includes NCOA2 and NCOA3, NCOA1 may interact with nuclear hormone receptors, including ESR, and other TFs, including AP1, c-Jun/c-Fos and HOXC11, as coactivators to facilitate the assembly of transcriptional protein complexes for chromatin remodeling and the activation of downstream target genes (27–31). Earlier studies identified an association between the increased expression of NCOA1 and enhanced angiogenesis, cell proliferation and survival, disease recurrence, a higher tumor grade and poor prognosis in a variety of cancer types. Analysis results from a low NCOA1 expression by our analysis results in this article appeared to contradict previous evidence; however, the GC dataset from Frycz et al (32) indicated that the mRNA expression of NCOA1 (P=0.00021) was significantly reduced in the tumoral mucosa compared with that in the adjacent healthy mucosa. Decreased levels of NCOA1 mRNA in GC tissue may be due to upregulation of cytochrome P450 family 19 subfamily A member 1 mRNA in the tumoral gastric mucosa, which causes dysregulation of two 17β-estradiol (E2) synthesis routes (the sulfatase and aromatase signaling pathways), resulting in E2 deficiency and inhibition of NCOA1 expression in E2-dependent methods (33). Furthermore, low expression of NCOA1 mRNA has been observed in bladder cancer urothelium samples (34), and the results of the present study were consistent with these results. NCOA1 not only acts as a coactivator, but has also been indicated to possess histone acetyltransferase activity. Sheppard et al (35) reported that the recruitment and induction of NCOA1 in the H3 sequence are each critical for the NCOA1-CBP interaction, which is necessary for ER function. Although the FC of NCOA1 expression was <2 in the GEO, TCGA and Oncomine datasets, the present results demonstrated that, no matter whether the expression of NCOA1 was upregulated or downregulated, aberrant expression of NCOA1 is significantly associated with poor prognosis and is an independent predictor for GC from our analysis (TCGA data: HR=0.639, 95% CI=0.437–0.933, P=0.020). This evidence suggested that weakly DEGs may also have important functions and roles in the occurrence and development of tumors. For instance, by quantifying the adenomatosis polyposis coli (APC) gene, Yan et al (36) determined that a weak reduction of APC expression was closely associated with the occurrence of hereditary colorectal tumors.
Unfortunately, no associations were observed between the aberrant expression of five specific genes (KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16) and clinical features when their potential clinical significance in patients with GC was explored in the TCGA dataset. Although age may serve as an independent prognostic factor according to the multivariate analysis, it was not a significant predictor for survival of patients whose genes were differentially expressed when stratified by age (data not presented). In addition, in a report on 97 patients with stage III–IVa/b head and neck squamous cell carcinoma, similar results were observed, in that NCOA1 expression was not significantly associated with any clinicopathological features, including sex, tumor site, T classification or nodal status (37). However, the majority of previous studies on certain malignant tumors indeed suggested that the aberrant expression of KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 was significantly associated with the patients' clinicopathological features (38,39). It is known that the molecular complexity and heterogeneity of GC made the elucidation of its specific pathogenesis challenging. All external or internal factors (e.g. poor dietary habits or genetic factors) and interwoven pathways may facilitate the occurrence of GC (1). Therefore, the heterogeneity between different studies on GC indicates a requirement for further large-scale studies to clarify and verify the data.
Histone modification, particularly acetylation and methylation, constitutes a vital epigenetic mechanism involved in the dynamic regulation of chromatin structure and gene expression (3,4). However, heterogenous factors associated with GC influence the onset and progression of histone modification reactions, including post-transcriptional regulation, the microenvironment, infection, immune responses and underlying genetic alterations. The upstream and downstream effects affect changes in the HMGs and associated enzymes that catalyze histone modifications, resulting in malignant processes. In the present study, five significant genes were identified: KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16, all of which have vital roles in histone modification. In addition to histone substrates, these enzymes may also directly catalyze non-histone substrates, including TFs, to perform target gene transcriptional regulation. KAT2A, encoding a histone lysine acetyltransferase, catalyzes the acetylation of lysine residues on histones, including H2B, H3 or H4 and TFs (40,41). PRDM16, a PR/SET domain family members, contains an N-terminal positive regulatory (PR) domain and zinc fingers, and functions as a histone 3 methyltransferase (42). Contributing to 90% of cellular PRMT activity and acting as a histone arginine methyltransferase, PRMT1 methylates histones, RNA-binding proteins and other TFs to epigenetically control the expression of downstream genes (43,44). SMYD5, a member of the histone lysine methyltransferase family, contains the catalytic SET domain and trimethylates H4K20, which has a critical role in carcinogenesis. However, due to the lack of a TPR domain in the structure, substrates of SMYD5 are limited on anything besides histones (45,46). Furthermore, it is worth mentioning that these genes also have important roles in drug resistance (47,48).
Underlying genetic alterations, including promoter methylation, copy number variations (CNVs) and chromosomal instability (CIN), have been reported to regulate histone modification-associated gene expression. For instance, low PRDM16 expression levels in non-small cell lung cancer and esophageal cancer were reported to be correlated with its promoter methylation (49,50). A high frequency of CNVs at 1p36.32 harboring the PRDM16 gene was observed in GC, suggesting that changes in gene CNVs also have vital roles in regulating gene expression (51). In addition, Burghel et al (52) indicated that PRDM16 was highly expressed in gained focal minimal common regions caused by CIN in colorectal cancer. Besides genetic variations, post-transcriptional regulations also affected the functioning of associated enzymes. For instance, NCOA1 was confirmed as a target of miR-223-3p and demonstrated to have low expression levels (53,54). Evidence has indicated that changes to the microenvironment resulting from hypoxia, which is insufficient to maintain cellular function, were associated with cancer pathology. In glioma cells, hypoxia decreased the gene expression of NCOA1 (55). It is known that Helicobacter pylori is responsible for gastric inflammation and gastric malignancy, which causes general inflammatory stress within the gastric mucosa, activating multiple oncogenic pathways and inducing epigenetic alterations, including histone modifications (56). These results suggested that factors upstream of histone modification-associated genes are important for the regulation of the expression of these genes.
Aberrant gene expression caused by upstream factors initiates cascade reactions, resulting in normal cells transforming into cancer cells and other types of malignant behavior. For instance, deletion of the KAT2A gene induces apoptosis in acute myeloid leukemia (57). Similarly, an increased apoptotic rate was observed in prostate cancer cells when PRDM16 expression was downregulated (58). Regarding cell regeneration and differentiation, histone modification-associated genes were also suggested to have important roles. PRMT1 methylates arginine on substrates Six1 or Eya1, and has been indicated to regulate muscle stem cell regeneration and differentiation (59). In embryonic stem cell differentiation, SMYD5 increases H4K20me3 and H3K9me3 levels and maintains chromosome integrity to ensure accurate differentiation (60). The functional basis of invasion and metastasis is the epithelial-to-mesenchymal transition (EMT). Previous studies have demonstrated that PRMT1, as a regulator, is closely associated with EMT. Katsuno et al (61) reported that PRMT1 is an essential mediator of transforming growth factor-r signaling, regulating the EMT and epithelial cell stemness by methylating SMAD7. Dysregulation of histone modification-associated genes not only promotes the acquirement of malignant biological phenotypes of cancer cells, but also has important effects on immunity. KAT2A, which is recruited by nuclear factor of activated T cells during the activation of T-cell receptor signaling pathways, methylates H3K9 of the interleukin-2 gene promoter to regulate T-cell activation and CD4+ T-cell differentiation into type 1 T-helper cells (Th1)/Th17 (62). In addition, methylation catalyzed by PRMT1 has been indicated to be required for pre-B-cell development and mature B-cell activation, together with B-cell translocation gene (63).
The present analysis on GC also indicated that, besides differential expression of NCOA1, our results on low PRDM16 and high PRMT1 levels were inconsistent with the corresponding contents of previous studies (64,65). Thee paradoxical results may be explained by the complexity of heterogeneous factors in GC. Furthermore, the varying expression profiles of KAT2A, NCOA1, SMYD5 and PRMT1 among different pathological classifications of gastric adenocarcinoma also demonstrated the presence of heterogeneity. A previous study by our group and other studies suggested that aberrant ATAD2 expression is associated with histone modification, hinting at a close correlation between ATAD2 and histone modification-associated genes (66). As speculated, the correlation and STRING interaction analysis indicated that ATAD2 may interact with ESR1 to regulate NCOA1 and PRMT1 in GC. Previous studies revealed that endogenous ATAD2 acts as a co-activator for ESR1 to activate downstream target gene expression, together with hormone-induced ESR1 recruiting to target genes at chromatin (67). NCOA1 also interacts with nuclear hormone receptors, including ESR1 (26). Of note, a negative correlation between NCOA1 and PRMT1 expression was observed in the present study, and the cross-talk among histone modification-associated genes was also previously reported (68). Certain limitations of the present study should be emphasized. Only a Bioinformatics analysis of external data was performed, and the expression of the five genes identified and interactions between NCOA1 and other genes, including the protein levels of ATAD2, were not verified through experimental methods as part of the present study. As a next step, verification in clinical samples from our center and assessment of potential interaction mechanism among ATAD2, ESR1 and NCOA1 will be performed. Hence, the results of such future experiments are to be anticipated.
In conclusion, the present study performed a comprehensive Bioinformatics analysis of the expression of histone modification-associated genes in GC and their association with prognosis. KAT2A, NCOA1, SMYD5, PRMT1 and PRDM16 were screened out and aberrant expression profiles were compared between GC and non-cancer tissues. Low NCOA1 expression was a closely associated with poor prognosis and was identified to be an independent predictor for GC. ATAD2 may interact with ESR1 to regulate NCOA1 and PRMT1 in GC. Due to the heterogeneity in GC, well-designed studies with larger sample sizes are required in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Science Foundation of Liaoning Province (grant no. 20170540567).
Availability of data and materials
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XM, JL, LW, TZ, XG, ZD and YZ analyzed the data. XM, ZZ and JL designed the study and prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Nagini S: Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 4:156–169. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Yang WY, Gu JL and Zhen TM: Recent advances of histone modification in gastric cancer. J Cancer Res Ther. 10 (Suppl):S240–S245. 2014. View Article : Google Scholar | |
|
Rice JC and Allis CD: Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol. 13:263–273. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A and Kirmizis A: N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet. 9:e10038052013. View Article : Google Scholar : PubMed/NCBI | |
|
Sakuraba K, Yokomizo K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H and Hibi K: TIP60 as a potential marker for the malignancy of gastric cancer. Anticancer Res. 31:77–79. 2011.PubMed/NCBI | |
|
He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL and Xie D: Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 13:3173–3178. 2013. View Article : Google Scholar | |
|
Goulet I, Gauvin G, Boisvenue S and Cote J: Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem. 282:33009–33021. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, et al: Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 66:11341–11347. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Hirst M and Marra MA: Epigenetics and human disease. Int J Biochem Cell Biol. 41:136–146. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Kang ZH, Wang CY, Zhang WL, Zhang JT, Yuan CH, Zhao PW, Lin YY, Hong S, Li CY and Wang L: Histone deacetylase HDAC4 promotes gastric cancer SGC-7901 cells progression via p21 repression. PLoS One. 9:e988942014. View Article : Google Scholar : PubMed/NCBI | |
|
Das C, Lucia MS, Hansen KC and Tyler JK: CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 459:113–117. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al: The polycomb group protein EZH2 directly controls DNA methylation. Nature. 439:871–874. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Na J, Lee K, Na W, Shin JY, Lee MJ, Yune TY, Lee HK, Jung HS, Kim WS and Ju BG: Histone H3K27 Demethylase JMJD3 in cooperation with NF-κB regulates keratinocyte wound healing. J Invest Dermatol. 136:847–858. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J and Dou Y: MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 30:5335–5347. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferré F, et al: The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 471:513–517. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Xiang Y, Zhu Z, Han G, Lin H, Xu L and Chen CD: JMJD3 is a histone H3K27 demethylase. Cell Res. 17:850–857. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng YC, Duan YC, Ma JL, Xu RM, Zi X, Lv WL, Wang MM, Ye XW, Zhu S, Mobley D, et al: Triazole-dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J Med Chem. 56:8543–8560. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and Varambally S: UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Gyorffy B, Surowiak P, Budczies J and Lanczky A: Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI | |
|
Wu G, Liu H, He H, Wang Y, Lu X, Yu Y, Xia S, Meng X and Liu Y: miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 14:1072014. View Article : Google Scholar : PubMed/NCBI | |
|
Venkatesh S and Workman JL: Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 16:178–189. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Audia JE and Campbell RM: Histone modifications and cancer. Cold Spring Harb Perspect Biol. 8:a0195212016. View Article : Google Scholar : PubMed/NCBI | |
|
Chan EM, Chan RJ, Comer EM, Goulet RJ III, Crean CD, Brown ZD, Fruehwald AM, Yang Z, Boswell HS, Nakshatri H and Gabig TG: MOZ and MOZ-CBP cooperate with NF-kappaB to activate transcription from NF-kappaB-dependent promoters. Exp Hematol. 35:1782–1792. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Yang XJ: The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32:959–976. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Rasti M, Grand RJ, Mymryk JS, Gallimore PH and Turnell AS: Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J Virol. 79:5594–5605. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Xu J, Wu RC and O'Malley BW: Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 9:615–630. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Fleming FJ, Hill AD, McDermott EW, O'Higgins NJ and Young LS: Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 89:375–383. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Qin L, Chen X, Wu Y, Feng Z, He T, Wang L, Liao L and Xu J: Steroid receptor coactivator-1 upregulates integrin α5 expression to promote breast cancer cell adhesion and migration. Cancer Res. 71:1742–1751. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Qin L, Liu Z, Chen H and Xu J: The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 69:3819–3827. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Gouon-Evans V, Rothenberg ME and Pollard JW: Postnatal mammary gland development requires macrophages and eosinophils. Development. 127:2269–2282. 2000.PubMed/NCBI | |
|
Frycz BA, Murawa D, Borejsza-Wysocki M, Wichtowski M, Spychala A, Marciniak R, Murawa P, Drews M and Jagodziński PP: mRNA expression of steroidogenic enzymes, steroid hormone receptors and their coregulators in gastric cancer. Oncol Lett. 13:3369–3378. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Tai H, Kubota N and Kato S: Involvement of nuclear receptor coactivator SRC-1 in estrogen-dependent cell growth of MCF-7 cells. Biochem Biophys Res Commun. 267:311–316. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Boorjian SA, Heemers HV, Frank I, Farmer SA, Schmidt LJ, Sebo TJ and Tindall DJ: Expression and significance of androgen receptor coactivators in urothelial carcinoma of the bladder. Endocr Relat Cancer. 16:123–137. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Sheppard HM, Harries JC, Hussain S, Bevan C and Heery DM: Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol. 21:39–50. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Yan H, Dobbie Z, Gruber SB, Markowitz S, Romans K, Giardiello FM, Kinzler KW and Vogelstein B: Small changes in expression affect predisposition to tumorigenesis. Nat Genet. 30:25–26. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Pavón MA, Parreño M, Téllez-Gabriel M, León X, Arroyo-Solera I, López M, Céspedes MV, Casanova I, Gallardo A, López-Pousa A, et al: CKMT1 and NCOA1 expression as a predictor of clinical outcome in patients with advanced-stage head and neck squamous cell carcinoma. Head Neck. 38 (Suppl 1):E1392–E1403. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Qin L, Wu YL, Toneff MJ, Li D, Liao L, Gao X, Bane FT, Tien JC, Xu Y, Feng Z, et al: NCOA1 directly targets M-CSF1 expression to promote breast cancer metastasis. Cancer Res. 74:3477–3488. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Gao H, Chakraborty G, Lee-Lim AP, Mavrakis KJ, Wendel HG and Giancotti FG: Forward genetic screens in mice uncover mediators and suppressors of metastatic reactivation. Proc Natl Acad Sci USA. 111:16532–16537. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K and Tora L: SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 44:410–423. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR and Roth SY: Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 26:229–232. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Sonnet M, Claus R, Becker N, Zucknick M, Petersen J, Lipka DB, Oakes CC, Andrulis M, Lier A, Milsom MD, et al: Early aberrant DNA methylation events in a mouse model of acute myeloid leukemia. Genome Med. 6:342014. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Zhu R, Wang W, Fu D, Hou J, Ji S, Chen B, Hu Z, Shao X, Yu X, et al: Arginine Methyltransferase 1 in the nucleus accumbens regulates behavioral effects of cocaine. J Neurosci. 35:12890–12902. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Boisvert FM, Rhie A, Richard S and Doherty AJ: The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 4:1834–1841. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Qian C and Zhou MM: SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 63:2755–2763. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D and Couture JF: Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 3:301–308. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Qu Y, Zhang XL, Xing J, Niu XL, Chen X and Li ZM: Autocrine production of interleukin-6 confers ovarian cancer cells resistance to tamoxifen via ER isoforms and SRC-1. Mol Cell Endocrinol. 382:791–803. 2012. View Article : Google Scholar | |
|
Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, et al: PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 125:4529–4543. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Tan SX, Hu RC, Xia Q, Tan YL, Liu JJ, Gan GX and Wang LL: The methylation profiles of PRDM promoters in non-small cell lung cancer. Onco Targets Ther. 11:2991–3002. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Peng X, Xue H, Lü L, Shi P and Wang J and Wang J: Accumulated promoter methylation as a potential biomarker for esophageal cancer. Oncotarget. 8:679–691. 2017.PubMed/NCBI | |
|
Bibi F, Ali I, Naseer MI, Ali Mohamoud HS, Yasir M, Alvi SA, Jiman-Fatani AA, Sawan A and Azhar EI: Detection of genetic alterations in gastric cancer patients from Saudi Arabia using comparative genomic hybridization (CGH). PLoS One. 13:e02025762018. View Article : Google Scholar : PubMed/NCBI | |
|
Burghel GJ, Lin WY, Whitehouse H, Brock I, Hammond D, Bury J, Stephenson Y, George R and Cox A: Identification of candidate driver genes in common focal chromosomal aberrations of microsatellite stable colorectal cancer. PLoS One. 8:e838592013. View Article : Google Scholar : PubMed/NCBI | |
|
Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L, et al: High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 8:11896–11905. 2017.PubMed/NCBI | |
|
Guo J, Cao R, Yu X, Xiao Z and Chen Z: MicroRNA-223-3p inhibits human bladder cancer cell migration and invasion. Tumour Biol. 39:10104283176916782017. View Article : Google Scholar : PubMed/NCBI | |
|
Minchenko OH, Garmash IA, Minchenko DO, Kuznetsova AY and Ratushna OO: Inhibition of IRE1 modifies hypoxic regulation of G6PD, GPI, TKT, TALDO1, PGLS and RPIA genes expression in U87 glioma cells. Ukr Biochem J. 89:38–49. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ding SZ, Goldberg JB and Hatakeyama M: Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 6:851–862. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, et al: A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17:1193–1205. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu S, Xu Y, Song M, Chen G, Wang H, Zhao Y, Wang Z and Li F: PRDM16 is associated with evasion of apoptosis by prostatic cancer cells according to RNA interference screening. Mol Med Rep. 14:3357–3361. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Blanc RS, Vogel G, Li X, Yu Z, Li S and Richard S: Arginine methylation by PRMT1 regulates muscle stem cell fate. Mol Cell Biol. 37(pii): e00457–16. 2017.PubMed/NCBI | |
|
Kidder BL, He R, Wangsa D, Padilla-Nash HM, Bernardo MM, Sheng S, Ried T and Zhao K: SMYD5 controls heterochromatin and chromosome integrity during embryonic stem cell differentiation. Cancer Res. 77:6729–6745. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Katsuno Y, Qin J, Oses-Prieto J, Wang H, Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame A, Xu J and Derynck R: Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J Biol Chem. 293:13059–13072. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Gao B, Kong Q, Zhang Y, Yun C, Dent SYR, Song J, Zhang DD, Wang Y, Li X and Fang D: The histone acetyltransferase Gcn5 positively regulates T Cell activation. J Immunol. 198:3927–3938. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wu GS and Bassing CH: Flip the switch: BTG2-PRMT1 protein complexes antagonize pre-B-cell proliferation to promote B-cell development. Cell Mol Immunol. 15:808–811. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Lei Q, Liu X, Fu H, Sun Y, Wang L, Xu G, Wang W, Yu Z, Liu C, Li P, et al: miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget. 7:5007–5022. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Altan B, Yokobori T, Ide M, Mochiki E, Toyomasu Y, Kogure N, Kimura A, Hara K, Bai T, Bao P, et al: Nuclear PRMT1 expression is associated with poor prognosis and chemosensitivity in gastric cancer patients. Gastric Cancer. 19:789–797. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wu G, Lu X, Wang Y, He H, Meng X, Xia S, Zhen K and Liu Y: Epigenetic high regulation of ATAD2 regulates the Hh pathway in human hepatocellular carcinoma. Int J Oncol. 45:351–361. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Zou JX, Revenko AS, Li LB, Gemo AT and Chen HW: ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci USA. 104:18067–18072. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K and Dent SY: Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis. Mol Cell Biol. 34:3746–3753. 2014. View Article : Google Scholar : PubMed/NCBI |