Digestive system infection by SARS‑CoV‑2: Entry mechanism, clinical symptoms and expression of major receptors (Review)
- Authors:
- Published online on: January 20, 2023 https://doi.org/10.3892/ijmm.2023.5222
- Article Number: 19
-
Copyright: © Zheng et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) is the pathogen responsible for causing SARS-CoV-2 pneumonia [coronavirus disease 2019 (COVID-19)]. The initial clinical symptoms of viral infection are frequently atypical and include mild coughing and headaches (1,2). The increase in SARS-CoV-2 infection cases has also led to the emergence of a number of characteristic stomach symptoms. According to a multicenter retrospective study by Rizvi et al (3), 3,229 (18.5%) of the 17,462 hospitalized patients exhibited various gastrointestinal manifestations. During follow-up, these manifestations included gastrointestinal bleeding, pancreatitis and conditions arising from the manifestations, such as malnutrition. Although the exact mechanism of SARS-CoV-2 infection in the gastrointestinal tract remains unknown, it is generally accepted that the expression of angiotensin-converting enzyme II (ACE2) and transmembrane serine protease II (TMPRSS2) in the stomach, liver, pancreas, duodenum and colon is essential for infection to occur (4). Through the tethering of the virus to both these proteins via its spike (S) protein, the virus is able to enter the digestive tract.
ACE2 is a vital part of the renin-angiotensin-aldosterone system, being produced from the conversion of ACE (5). ACE2 acts as a key effector peptide that causes vasodilation (5). TMPRSS2 is a type II transmembrane protein that acts as a serine protease (6,7). The principal mediators of viral S protein binding are these two proteins. Several factors, such as age, sex, obesity, smoking, Helicobacter pylori (H. pylori) infection and tumors, have been indicated to be capable of promoting the upregulation of the genes encoding ACE2 and TMPRSS2 in the digestive system, making it easier for SARS-CoV-2 to enter the body (5,7). For instance, two recent studies revealed that stomach and colorectal adenocarcinomas have high levels of ACE2 and TMPRSS2 expression (8,9). In addition, Viveiros et al (10) reported that older male mice expressed higher levels of ACE2 compared with younger mice. Furthermore, Da Eira et al (11) observed that obese mice have comparatively higher levels of ACE2 and TMPRSS2 expression.
Taken together, the findings of previous studies have established that the entry of SARS-CoV-2 into the digestive tract, which causes diarrhea, nausea, vomiting and lack of appetite, depends on ACE2 and TMPRSS2. In addition, SARS-CoV-2 is able to influence the severity, prognosis and outcome of conditions such as liver damage and severe pancreatitis. It also has a direct or indirect association with a number of common digestive ailments. Through introducing the microstructure of SARS-CoV-2 and detailing the process via which SARS-CoV-2 enters the human body through ACE2 and TMPRSS2, the present review summarizes the effects of SARS-CoV-2 infection on the digestive system and the expression characteristics of ACE2 and TMPRSS2 in major target organs. Several potential strategies for the diagnosis, identification and treatment of digestive system diseases during the COVID-19 pandemic are also described.
2. Structural features and molecular mechanism of SARS-CoV-2 entry into cells
Classification and microstructure of SARS-CoV-2
At present, coronavirus has been divided into four major subclades, namely the α-, β-, γ- and δ-coronaviruses. SARS-CoV-2 belongs to the β-coronavirus group and mainly arises in humans and mammals. It belongs to the same branch of coronaviruses as SARS-CoV, SARS, human coronavirus (HCoV)-OC43, HCoV-HKU1 and Middle East Respiratory Syndrome coronavirus (MERS-CoV) (6,12,13). SARS-CoV-2 and SARS-like coronavirus share 88% homology, whereas SARS-CoV-2 and SARS-CoV share 79% homology, and SARS-CoV-2 and MERS-CoV share a relatively low homology of 50%. However, a computational analysis of the crystal structure of SARS-CoV-2 revealed that its manner of attachment to host ACE2 was comparable with that of the SARS-CoV and HCoV-NL63 viruses (13).
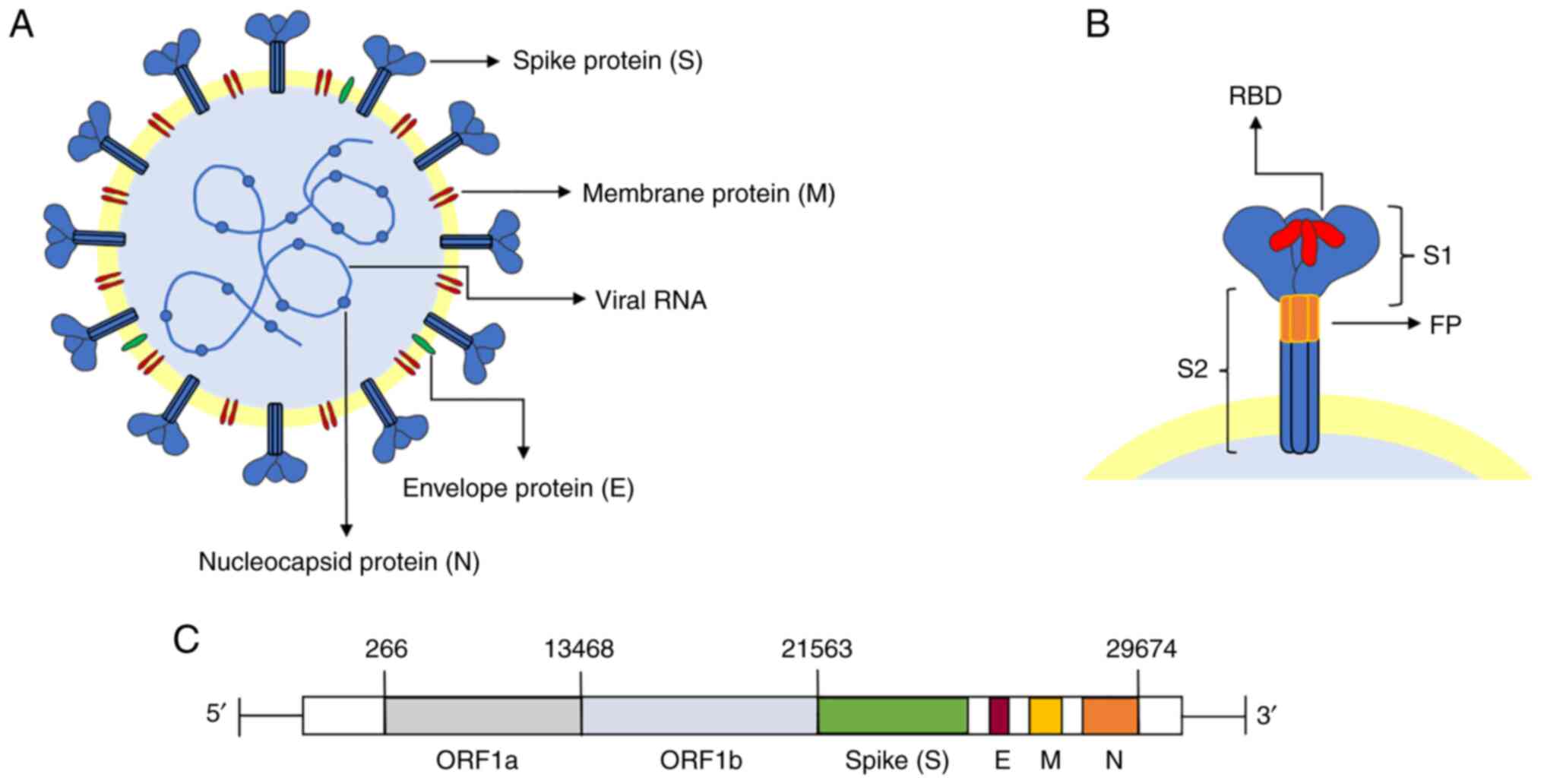
The viral particles of SARS-CoV-2 are single-stranded positive RNA envelope particles, having a spherical shape and a size between 100 and 160 nm. Its genome, 29,903 bp in length, has been demonstrated to have a highly conserved structure (12,14,15). The genome includes three regions. The open reading frames (ORFs) ORF1a and ORF1b comprise the first two-thirds of the genome, whereas the final third of the genome encodes structural proteins, including the S protein, envelope (E) protein, membrane (M) protein and nucleocapsid (N) protein (Fig. 1) (12).
The S protein, one of the four abovementioned structural proteins, is essential for viral entrance into the host cells. The S protein is a homotrimeric class I membrane protein (6,16). Located on the surface of the virus particles, the protein forms a crown (17) and, indeed, SARS-CoV-2 acquired its name for this reason (Fig. 1B). There are 22 N-linked glycosylation sites on each monomer of the highly glycosylated protein (66 in total) (18). The S protein contains an N-terminal signal peptide, the S1 subunit (responsible for receptor binding) and the S2 subunit (responsible for membrane fusion), with a total length of 1,273 amino acid residues (18). Receptor binding is performed by the S1 (N-terminal) subunit, whereas membrane fusion is accomplished by the S2 (C-terminal) subunit (16). The N-terminal domain, receptor-binding domain (RBD) and C-terminal domain 1 (CTD1) and CTD2 are other divisions of the S1 subunit. Regarding the S2 subunit, this comprises a central helix, junction region, heptapeptide repeat sequence 1 (HR1) and HR2, fusion peptide (FP), FP proximal region, transmembrane fragment and cytoplasmic tail (18). According to recent studies, RBD and FP are the structures specifically linked to viral invasion.
Molecular mechanism through which SARS-CoV-2 enters the human body
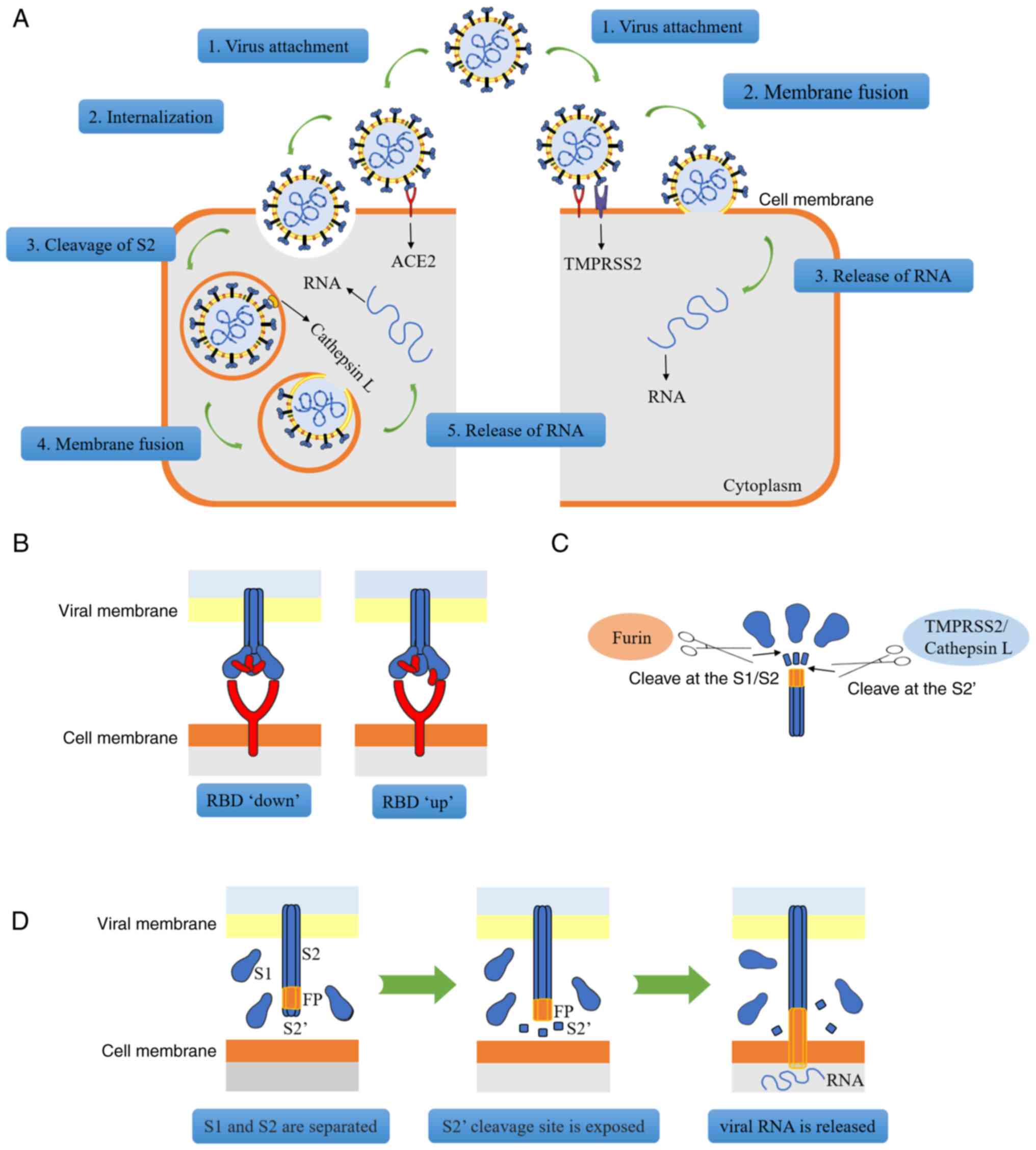
The RBD, a protein mainly composed of four cysteine residues that form disulfide bonds and seven β-fragments, is able to bind to ACE2 (17,19). Viruses may enter host cells in two different ways (Fig. 2A). In the first scenario, if the host cell only expresses ACE2 but does not express TMPRSS2 (or if the expression of TMPRSS2 is insufficient), the viral S protein may bind to ACE2 to change the conformation of the S1 subunit, thereby exposing the S2 subunit; subsequently, via the process of reticulin-mediated endocytosis, the entire viral molecule enters the cell and after the S2 subunit is dissociated by cathepsins, channels are opened for the release of viral RNA (6). Alternatively, if the host cell co-expresses ACE2 and TMPRSS2, viral RNA enters the cell through membrane fusion. First, ACE2 combines with RBD, resulting in a change in the conformation of the S protein (Fig. 2B). At this stage, the furin protease, a proprotein conversion enzyme that is able to activate the S protein, recognizes and cleaves the polybase insertion (also termed 'PRRAR') site in S1/S2, and S1/S2 subunit dissociation occurs (20,21). After S1 is separated from S2, the conformation of the S2 subunit undergoes a change, thereby exposing the S2' cleavage site, which is recognized and cleaved by TMPRSS2 (or cathepsin B/L), and then the exposed fusion peptide is inserted into the target cell membrane (12,22). Subsequently, HR1 and HR2 of the S2 subunit form a stable six-helix bundle fusion core that binds the viral and cellular membranes together to form a fusion pore to release the RNA (Fig. 2C and D) (15).
Previous studies have also indicated that mutant strains may enhance their ability to bind to ACE2 due to their own mutations, and for other reasons. For instance, the B.1.1.7, B.1.351, P.1 and B.1.617.2 mutant variants of SARS-CoV-2 have a high affinity for ACE2 and these mutant strains express multiple mutations of the S protein. These mutations may change the structure of the protein, resulting in greater infectivity (23-26). Storti et al (27) discovered that the B.1.1.7 strain is able to internalize faster and this accelerated internalization may be directly associated with the N501Y mutation of the S protein, which enhances the binding of RBD to ACE2. When the RBD gene is mutated, it may lead to an increase in the infection rate of SARS-CoV-2 (28). For instance, the D164G mutation was indicated to change the structure of the S protein, thereby increasing the affinity between RBD and ACE2 (29). Although limited studies have been published on the infection of the digestive system by mutant strains of SARS-CoV-2, the potential threat cannot be denied.
3. Clinical manifestations of SARS-CoV-2 infection and gastrointestinal expression of ACE2 and TMPRSS2
Clinical features of SARS-CoV-2 that damage the digestive system
SARS-CoV-2 causes atypical gastrointestinal symptoms, including bleeding
Since SARS-CoV-2 swept across the globe, a sizable number of digestive symptoms or disorders have been linked to viruses, primarily the most widespread and fundamental of digestive symptoms. SARS-CoV-2 has been positively identified in stool tests of numerous patients with COVID-19, which may provide evidence of the replication and presence of the virus in the digestive tract (30). In another SARS-CoV-2 RNA sample test, 44 (29%) of 153 stool samples were found to be positive for SARS-CoV-2 (31). Certain patients with COVID-19 have tested positive for the virus in their stool, even after throat swab tests were negative (32,33). In addition, SARS-CoV-2 RNA was found in the stool of 53% of hospitalized patients with COVID-19, and biopsies of the stomach, duodenum and rectum tested positive for the viral nucleocapsid, suggesting that the virus may infect the digestive tract (34). Xiao et al (35) performed a gastrointestinal endoscopy on a patient with COVID-19 and found that the esophageal mucosa was damaged; a subsequent histological examination revealed the presence of a large number of plasma cells and lymphocyte infiltration in the lamina propria of the stomach, duodenum and rectum. In addition, viral nucleocapsid proteins were detected in the cytoplasm of these sites. The prevalence of gastrointestinal symptoms in individuals with COVID-19 ranges from 12 to 61%, and gastrointestinal symptoms were manifested in the digestive tract more frequently in patients with a longer disease course (36). The signs and symptoms include diarrhea, nausea and vomiting. Diarrhea is the most typical symptom (37). Retrospective case studies have indicated that diarrhea, nausea, vomiting and anorexia are the most typical digestive symptoms in patients with COVID-19 (38-40). In addition, evidence suggests that nausea frequently starts at an early stage in patients infected with SARS-CoV-2, indicating that the gastrointestinal tract may be infected by the virus. Nausea was associated with the first case of COVID-19 in both China and the USA (41). In excess of 12,000 patients with COVID-19 were included in the analysis of 41 studies by Andrews et al (42), and the findings revealed a median incidence of nausea and diarrhea of 10.5 and 11%, respectively. Therefore, patients with SARS-CoV-2 infection should be vigilant of symptoms of both nausea and diarrhea with similar care. Epidemiological data have also indicated that nausea should be taken into consideration as a potential early symptom of SARS-CoV-2 (41). The most frequent abdomen computed tomography findings for imaging viral invasion of the digestive tract in patients with COVID-19 were found to be colonic thickening and edema, gastritis and small intestine gas buildup (43).
Several SARS-CoV-2-positive individuals have also experienced gastrointestinal hemorrhage. Carvalho et al (44) reported on a 71-year-old female patient with hemorrhagic colitis brought to the hospital on account of SARS-CoV-2 infection; endoscopic investigation identified a patchy localized erythema without any ulcer in the gut. The lamina propria had modest enlargement and edema, as observed by H&E staining of the colon and rectal biopsy. In addition, the particular case of a 77-year-old male patient with upper gastrointestinal hemorrhage was reported by Li et al (45). Lymphocyte infiltration and the presence of SARS-CoV-2 RNA in the samples taken from this patient provided conclusive evidence that the upper gastrointestinal bleeding in this case was caused by esophageal SARS-CoV-2 infection. A total of 95 patients with COVID-19 were studied by Lin et al (38), six of whom received endoscopies due to gastrointestinal symptoms. A severely ill patient also experienced esophageal hemorrhage, erosion and ulceration. In addition, SARS-CoV-2 RNA was found in the rectum, duodenum, stomach and esophagus of two seriously ill patients.
In the event of an epidemic breakout, symptoms that may be encountered in clinical practice include nausea, diarrhea and stomach pain. It is fitting to contemplate the connection between these manifestations and SARS-CoV-2, if other possible causes that may account for the gastrointestinal symptoms have been ruled out. For hospitalized patients, it is also important to look out for SARS-CoV-2 infection to prevent the virus's impact on the onset and prognosis of digestive illnesses.
Effects of SARS-CoV-2 on the digestive system may be prolonged
Hospitalization of patients with COVID-19 with SARS-CoV-2 gastrointestinal infections led to a prolongation of the symptoms in 104 patients in China, according to a retrospective cohort research study (46). Hu et al (47) tested for the virus in 289 patients with COVID-19 and 21 (7.3%) of these patients were readmitted after discharge due to re-detection of SARS-CoV-2. In the positive retest, the percentage of the readmitted patients for whom the virus was detected by anal swab was 71.4% (15/21). The subsequent phylogenetic analysis of the patients' full-length SARS-CoV-2 genome revealed that the virus detected in the retest had evolved from the parental virus involved in the initial infection. Therefore, the virus was indicated to participate in the primary, rather than the secondary infection. Hu et al (47) also determined that the presence of SARS-CoV-2 may remain undetected and that the virus may be replicated at low levels, mainly in the gastrointestinal tract, for a long time; furthermore, periodic shedding of the virus may lead to a resurgence of the virus, as mainly found in the anal swab samples. In patients with acute diarrhea, Noviello et al (48) discovered that moderate gastrointestinal symptoms persisted for ~5 months after SARS-CoV-2 infection. In addition, these researchers considered that acute SARS-CoV-2 infection may also have an impact on the brain-gut axis, resulting in symptoms such as headaches, backaches, irregular sleep patterns, low mood and anxiety (48).
Furthermore, patients with underlying gastrointestinal problems may potentially be impacted by SARS-CoV-2. In a retrospective analysis of surgical resection specimens from patients with gastrointestinal cancer, the specimens were tested for SARSCoV-2 to determine whether the patients had COVID-19. The results indicated that among 52 patients with gastrointestinal cancer, the mortality rate of patients with early or asymptomatic COVID-19 following surgery was 16.7%, which was much higher compared with that of patients without COVID-19 (49). In addition, the presence of certain underlying conditions, such as inflammatory bowel disease (IBD), was indicated to boost the expression of ACE2, with the result that the patients may have been more susceptible to SARS-CoV-2 (50). There is evidence that patients with IBD express ACE2 and TMPRSS2 more frequently in the colorectum compared with non-IBD patients (51,52). The study by Tao et al (53) revealed that patients with COVID-19 with IBD were more likely to experience symptoms of diarrhea and abdominal pain, and have elevated levels of biomarkers compared with patients without IBD. Due to viral infection, colitis is easily induced via direct damage caused to the intestinal epithelial cells. As a result, patients with IBD may be more vulnerable to intestinal damage caused by COVID-19 (54). Viganò et al (55) surveyed 709 patients with IBD, 53 of whom were also infected with COVID-19. They found that the probability of diarrhea was 49%, significantly higher than the probability for patients with IBD alone (42.2%). Furthermore, active IBD has been indicated to be significantly associated with COVID-19. Derikx et al (56) also found that ~38.6% of patients with IBD who were infected with COVID-19 had diarrhea symptoms. IBD patients with COVID-19 are also more likely to develop digestive diseases than those infected with COVID-19 or IBD alone, suggesting that COVID-19 may be associated either with the aggravation of IBD symptoms or with the promotion of its transition to the active phase.
The gastrointestinal tract may become infected with SARS-CoV-2 on a recurring, periodic and persistent basis. Patients with COVID-19 must continue to be monitored in order to assess whether the virus is still active after their gastrointestinal symptoms have improved. In addition, as far as possible, SARS-CoV-2 infection should always be avoided by those with fundamental digestive problems, in order to prevent the condition from getting worse or changing the prognosis.
SARS-CoV-2 with TMPRSS2 and ACE2 expression in the gastrointestinal tract
Connection between ACE2, SARS-CoV-2 and the stomach
Both healthy individuals and SARS-CoV-2-infected patients have digestive organs that express ACE2 and TMPRSS2. ACE2 is expressed in stomach tissues, according to recent investigations (57,58). According to Lee et al (59), who performed single-cell RNA sequencing (scRNA-seq) analyses of various parts of the gastrointestinal tract, the expression ratios of ACE2 in the upper digestive tract (esophagus, stomach and duodenum) and the lower digestive tract (ileum and colorectum) were 1.04% (1,084/104,174 cells) and 14.06% (4,754/33,808 cells), respectively. In the gastrointestinal tract, the co-expression of ACE2 and TMPRSS2 was found to be the highest in the small intestine and colorectum. More than 20% of intestinal epithelial cells and ~5% of colon cells were found to co-express ACE2 and TMPRSS2 (59). According to An et al (60), the main cells of the stomach manufacture pepsinogen and express ACE2 at a higher level in gastric tissue than in parietal cells, which may help to explain why patients with SARS-CoV-2 who have anorexia display this clinical characteristic. Due to the fact that ACE2 is still expressed in the gastric mucosa, further investigations have discovered that H. pylori infection and intestinal metaplasia may render subjects vulnerable to SARS-CoV-2 (61). Finally, Sun et al (62) developed a human ACE2 (hACE2)-expressing mouse model and demonstrated that hACE2 mice are sensitive to SARS-CoV-2 infection.
Characteristics of SARS-CoV-2 intestinal infection and expression of ACE2 and TMPRSS2 in intestinal cells
Numerous studies have confirmed that the intestinal tract is one of the susceptible sites for SARS-CoV-2. SARS-CoV-2-infected gastrointestinal tracts were indicated to shed a large number of infectious viruses, according to an African green monkey viral infection experiment. Furthermore, infectious viruses were repeatedly isolated over time from mucosal swabs, including from the rectum (63). Using monkey experiments, Jiao et al (64) also indicated that SARS-COV-2 infection led to an inhibition of gastrointestinal goblet cell proliferation, with the induction of apoptosis. Goblet cells secrete mucins, which prevent the entry of pathogens into the cells; a lack of mucins renders hosts more susceptible to pathogens. Therefore, SARS-CoV-2 infection has been shown to result in an inhibition of the proliferation of goblet cells and a decrease in the levels of mucins, which induces apoptosis, leading to the destruction of the intestinal barrier and further infection of multiple tissues (64).
Regarding the human gut, Livanos et al (65) observed strong expression of ACE2 on the brush border of the small intestine in both uninfected and SARS-CoV-2-infected patients. In addition, using immunofluorescence and electron microscopic experiments, these researchers detected viral nucleocapsid proteins in the small intestinal epithelial cells of 11 out of 12 COVID-19 patients (mainly goblet cells), suggesting that these cells were infected with the virus. In addition, Lee et al (59) found that ACE2 and TMPRSS2 are mainly co-expressed in the intestinal epithelial cells of the lower digestive tract, with the highest co-expression rates occurring in progenitor and stem-like epithelial cells, particularly in the small intestine, suggesting a potential mechanism for the gastrointestinal manifestations of acute COVID-19 infection. As far as the large intestine is concerned, studies have confirmed that ACE2 is mainly expressed on the membrane and in the cytoplasm of goblet cells, and the expression of ACE2 on the basal side of the colonic epithelium was found to be lower than that on the luminal side. Since colonic basal cells are able to regenerate and differentiate into mature functional glandular epithelial cells, the damaged colonic epithelial cells will be repaired after the SARS-CoV-2 virus is cleared, which may provide the explanation for the self-limiting diarrhea found in patients with COVID-19 (60). Qi et al (66) analyzed scRNA-seq data from the digestive system and found that ACE2 and TMPRSS2 are highly expressed in goblet cells throughout the intestine (both small intestine and large intestine), thereby identifying the intestine as a high-risk organ.
A large number of previously published studies (60,65) have otherwise demonstrated that ACE2 and TMPRSS2 are fully expressed in numerous intestinal regions, particularly in goblet cells. The severity of a viral infection in the intestine may be directly correlated with the expression of these two proteases. Consequently, close attention should be paid to the areas where these proteases are highly expressed while a clinical diagnosis is being made.
4. SARS-CoV-2 and the liver
General characteristics of liver damage with SARS-CoV-2
According to the study by Wang et al (67), 303 (46.1%) of 657 patients with COVID-19 experienced liver injury, with the frequency of severe and critical cases being greater [148/257 (57.6%)] compared with that of moderate cases [155/400 (38.8%)]. Males [192/303 (63.4%)] were also found to be substantially more likely to experience liver damage [111/303 (36.6%)] as compared to females. Zhang et al (68) discovered that male patients were more likely to experience liver injury than female patients in a multivariate analysis of 218 patients performed at Wuhan Central Hospital. This increased likelihood of liver injury in male patients may have been due to the liver-protecting effects of increased estrogen levels in women. High levels of D-dimer and neutrophils are additional risk factors for liver damage in individuals with COVID-19, in addition to their sex. The prognosis may be poor for individuals who have underlying liver illness, such as alcoholic liver disease or cirrhosis, if they contract SARS-CoV-2 (69). Yang et al (70) noted in in vivo studies that the livers of hamsters infected with SARS-CoV-2 displayed structural abnormalities and had large vacuoles. The expression of the viral protein was compatible with the placement of large vacuoles in hepatocytes, as revealed by immunohistochemical nucleoprotein staining, indicating viral replication in the liver. A previous study also revealed that patients with COVID-19 who had liver injury had a high viral load during the early stages of infection, suggesting that the virus may harm the liver directly (71). When SARS-CoV-2 infects cells, a huge number of inflammatory mediator and chemokine molecules are released, which leads to the aggregation of neutrophils. In addition, their secretions, containing cytokines and chemokines, encourage immune cell aggregation and the immune response (72). According to another previously published study, the inflammatory cytokine storm that SARS-CoV-2 generates raises patients' levels of interleukin (IL)-1β, IL-2, IL-6, IL-8, IL-10, IL-17, interferon, interferon-γ-induced protein 10 and monocyte and results in damage to the liver (73).
The prognosis is typically poor once a patient is infected with SARS-CoV-2, whether or not there is an underlying liver condition. The liver is more susceptible to injury in males due to their levels of estrogen being lower than those in females. Therefore, when male patients are infected with SARS-CoV-2, more focus should be placed on liver protection.
SARS-CoV-2 infection is able to cause liver damage that may result in increased liver enzyme levels
In a large tertiary care health system in Detroit (USA), out of 1,935 patients who were hospitalized with COVID-19, 1,031 (53.2%) of them had mildly elevated levels of liver enzymes and 396 (20.5%) had liver damage (74). According to a meta-analysis study performed by Wijarnpreecha et al (75), a quarter of all patients with COVID-19 had increased levels of liver enzymes and the extents of the increases were associated with the severity of the condition. Previous studies have also demonstrated that liver damage, which manifests as high levels of the enzymes alanine aminotransferase (ALT), aspartate aminotransferase (AST), lactate dehydrogenase and total bilirubin, was observed in numerous patients with COVID-19, particularly in those who were critically ill. Typically, the levels peak in 8-9 days, whereas in patients only moderately affected by COVID-19, the increase in these indicators was barely perceptible (41,76,77). According to a retrospective study by Gomi et al (78) that included 216 subjects, patients with mild and moderate COVID-19 infection were more likely to have impaired liver function. In addition, it is possible to distinguish COVID-19 from other illnesses using elevated ALT and AST without alkaline phosphatase or γ-glutamyl transpeptidase. These studies, in addition to others, have found that it is the activation of hepatic infiltrating lymphocytes, which results in the rise of hepatic cytokine levels, that causes the indirect liver harm resulting from SARS-CoV-2 infection.
Types and multiple factors of liver injury induced by SARS-CoV-2
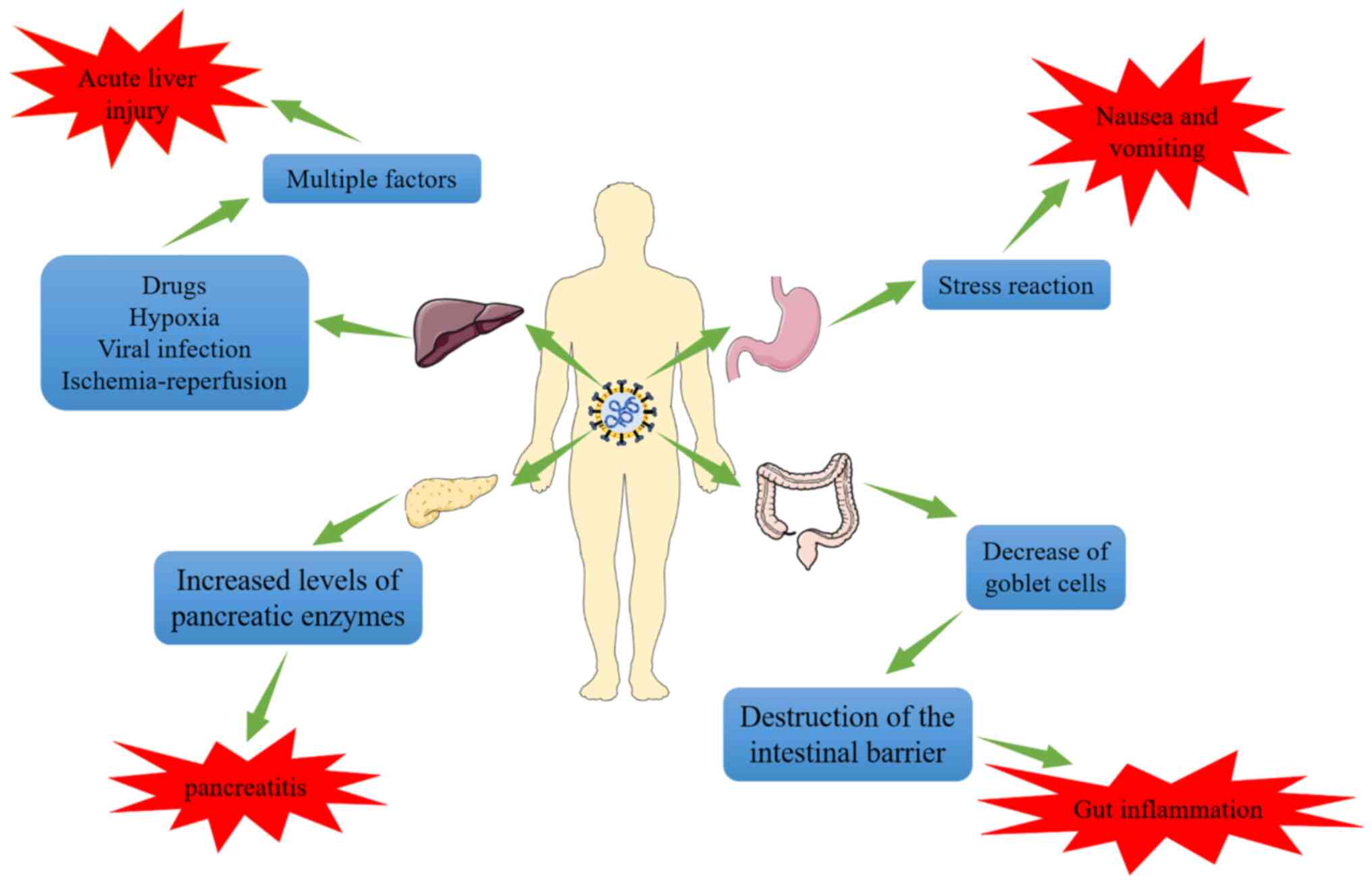
There is evidence that liver injury in patients with COVID-19 is not only caused by SARS-CoV-2, but also that SARS-CoV-2 infection is associated with drug-induced liver injury, secondary liver injury caused by hypoxia, viral liver injury and liver ischemia-reperfusion injury (Fig. 3) (73,79,80). Studies have indicated that SARS-CoV-2 may infect endothelial cells and cause diffuse dermatitis. Subsequently, microvascular dysfunction may lead to hypercoagulable states, tissue edema and organ ischemia (81,82). In addition, a pathological examination of one case of mortality resulting from COVID-19 revealed moderate microvascular steatosis and mild active inflammation of the hepatic lobule-portal vein region in the liver, which indicated that the liver injury of COVID-19 is frequently secondary damage caused by hypoxia (83). In vitro experiments also suggested that the expression and activity of ACE2 increased markedly in hepatocytes and cholangiocytes under hypoxic conditions (84). A retrospective analysis of 551 patients with COVID-19 in New York (USA) examined liver function during the time that they were hospitalized, and this analysis revealed that SARS-CoV-2 and other factors may have contributed to liver injury (85). According to the study by Chew et al (86), anomalies in liver tests linked with COVID-19 were predominantly found to be secondary to ischemia or drug-induced liver injury. As localized necrosis and liver neutrophil and Kupffer cell growth were observed in the autopsies of individuals who had died from COVID-19, Vishwajeet et al (87) concluded that SARS-CoV-2 and other factors probably contribute to liver damage in patients with COVID-19. Del Nonno et al (77) discovered from liver biopsies from three patients with coronavirus pneumonia and the autopsies of three individuals infected with SARS-CoV-2 that in an increase in the amount of iron in the liver. It is hypothesized that SARS-CoV-2 may elevate serum ferritin levels and cause damage to the liver. Multifactorial liver injury, however, has increased the difficulty of treating liver injury during the SARS-CoV-2 pandemic.
5. SARS-CoV-2-infected pancreatic symptoms and expression of associated proteins
Acute pancreatitis may be caused by SARS-CoV-2
Acute pancreatitis, and even abrupt onset diabetes with ketoacidosis, are common in adult patients with COVID-19, and numerous case reports over the course of the last two years have demonstrated that SARS-CoV-2 appears to be able to affect pancreatic (exocrine and endocrine) cells (88-90). Elevated lipase or amylase levels, or other potential explanations, may be eliminated when patients with COVID-19 experience abdominal pain and do not also have any underlying digestive illnesses. In a case study involving three family members, the mother and daughter both experienced severe acute pancreatitis (thereby eliminating other potential causes of pancreatitis) and had increased levels of pancreatic lipase (91). In a different cohort study, 83 patients hospitalized with COVID-19 also exhibited increased lipase levels (>3 times the upper limit of normal), which were regarded as a separate indicator of severe illness (92). According to a retrospective cohort analysis, out of 48,012 hospitalized patients, 11,883 (24.75%) tested positive for SARS-CoV-2 at admission and 189 (point prevalence, 0.39%) met the diagnostic criteria for pancreatitis. Of these 189 individuals, 32 (17%) tested positive for SARS-CoV-2. The point prevalence of pancreatitis among COVID-19 hospitalized patients was found to be 0.27% (93). Children have also been reported to have acute pancreatitis. According to a case report by Samies et al (94), three children who were hospitalized for severe pancreatitis also tested positive for SARS-CoV-2 in blood tests. Although pancreatitis is not a direct result of SARS-CoV-2 infection, there appears to be an association between pancreatitis and the COVID-19 diagnostic timeframe. In addition, subjects who already have pancreatitis may be affected by SARS-CoV-2. In a multicenter investigation, Pandanaboyana et al (95) discovered that, among 1,777 individuals with acute pancreatitis, the incidences of severe pancreatitis, morbidity and death were significantly higher when SARS-CoV-2 infection was present.
SARS-CoV-2 may induce acute pancreatitis, or is a risk factor for acute pancreatitis, impacting its severity and prognosis, even though the connection between acute pancreatitis and SARS-CoV-2 infection is still being investigated.
Correlation between SARS-CoV-2, ACE2, TMPRSS2 and pancreatic infection
Pancreatic cell expression of SARS-CoV-2-associated proteins
It has been reported that ACE2 is expressed in pancreatic exocrine and endocrine tissues (96). Co-expression of ACE2 and TMPRSS2 in pancreatic duct and acinar cells has been demonstrated in numerous studies to be essential for effective viral entry into cells. Pancreatic damage is induced by viral attachment to ACE2 (59,97,98). In a previous study, researchers examined the expression of TMPRSS2 in 30 normal human tissues and discovered that the target organs of SARS-CoV-2 infection, namely the stomach, pancreas, lung, small intestine and salivary glands, had the highest levels of TMPRSS2 expression (99). This protease is considered to be the primary serine protease required for SARS-CoV-2 infection, since studies have revealed that TMPRSS2 is expressed at a relatively higher level in duct and acinar cells compared with β-cells (100). Previous studies also indicated that TMPRSS2 is primarily expressed in duct cells, although ACE2 and TMPRSS2 are only infrequently co-expressed in pancreatic duct and endocrine cells. ACE2 is primarily expressed in islets and exocrine tissue capillaries, including pericytes and a subset of duct cells (59,101). An et al (60) used immunohistochemistry staining to identify the expression levels of the ACE2 protein in the colon, stomach, liver and pancreas. This group discovered that islet cells were stained with ACE2 more strongly than acinar cells. In addition, it has also been previously demonstrated that SARS-CoV-2 is able to enter islet cells and infect them, resulting in diabetes, which may be attributed to the selective expression of the cell-surface receptors neuropilin-1 (NRP1) and transferrin receptor (TFRC) in cells (22). According to this study, the enhancement in the levels of NRP1 and TFRC may be a potential mechanism for the SARS-CoV-2 tropism of cells.
SARS-CoV-2-induced pancreatic injury-associated protein phenotypes
In the islets of six of 11 patients with COVID-19, Steenblock et al (102) employed RNA in situ hybridization to identify the RNA of the virus SARS-CoV-2. Another study (103) identified that SARS-CoV-2 may penetrate and infect induced pluripotent stem cell-derived pancreatic cells, including endocrine and exocrine cell types. This resulted in morphological abnormalities and impaired expression of critical markers, which corresponds to inflammatory features. In addition, the postmortem pancreas examination of a patient who died from SARS-CoV-2-19 revealed SARS-CoV-2-19 infection of the pancreatic tissue. Therefore, taken together, these findings suggest that pancreatic cells may be directly infected by SARS-CoV-2. Drug-induced pancreatic injury cannot be ruled out; however, certain patients with digestive issues who are treated in hospitals also have a history of medicine use (96). According to data from one study, ACE2 is not expressed in pancreatic acinar cells, but only in islets and duct cells (60). 83.6% (56/67) of the biopsy islet tissues were found to be labeled positively for ACE2, whereas 15.3% (20/131) of the biopsy pancreatic acinar cells were mildly stained for ACE2 (60). In addition, capillary endothelial cells on 98.5% (129/131) of the acinar cells stained positive for ACE2. It is possible that SARS-CoV-2 may spread through the duct system cells in pancreatic tissue, eventually causing islet destruction and aberrant blood glucose levels. Therefore, this phenotype would result from the unique expression of SARS-CoV-2 in the pancreas following infection. Consequently, SARS-CoV-2 infection, and the ensuing damage, may increase due to the elevated expression level of ACE2 in the pancreas (60). According to other studies, SARS-CoV-2 infection and pancreatic cell inflammation may activate pancreatic stellate cells and cause fibrosis, as seen in infected non-human primate and human pancreas (97). As a result, ACE2 expression is essential for pancreatic SARS-CoV-2 infection.
6. Conclusions
Investigations are ongoing to determine how SARS-CoV-2 infects the gastrointestinal tract. It is known that the SARS-CoV-2 S protein binds to ACE2 of the host cell, cleaves the S protein with the help of proteases such as TMPRSS2 and then forms fusion pores to release RNA into the cytoplasm. The virus is multiplied in infected cells, which sets off an inflammatory reaction. It is not possible to rule out the possibility of SARS-CoV-2 infection when diagnosing digestive illnesses in the setting of the outbreak. The risk of SARS-CoV-2 cannot be denied, regardless of whether it directly affects the target organ, causing severe pancreatitis, gastrointestinal hemorrhage or liver damage, or whether it indirectly aggravates these conditions. Secondly, it is important to take into account both drug and viral harm to target organs while treating digestive system disorders, and to minimize the combined effects of medications. It is possible to investigate how to lessen the injury caused by SARS-CoV-2 to the digestive system by moderately decreasing the expression of ACE2 and TMPRSS2, or by creating medications that block the viral S protein. If there are further entry points into the digestive system, this will require extensive testing. Finally, the damage that SARS-CoV-2 causes to the digestive system should always be a concern.
Availability of data and materials
Not applicable.
Authors' contributions
LZhe and LZha drafted the manuscript and contributed equally. YZ participated in the literature search and analysis of the data to be included in the review. JA and HJ were involved in the design of the study and assisted in the preparation of the figures and tables. GW and BT edited and revised the manuscript. All authors have read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 82073087 and 81960507) and the Collaborative Innovation Center of Chinese Ministry of Education (grant no. 2020-39).
References
|
Li J, Li C, Wang X, Wang Y and Zhou Y: Considerations and perspectives on digestive diseases during the COVID-19 pandemic: A narrative review. Ann Palliat Med. 10:4858–4867. 2021. View Article : Google Scholar | |
|
Delgado-Gonzalez P, Gonzalez-Villarreal CA, Roacho-Perez JA, Quiroz-Reyes AG, Islas JF, Delgado-Gallegos JL, Arellanos-Soto D, Galan-Huerta KA and Garza-Treviño EN: Inflammatory effect on the gastrointestinal system associated with COVID-19. World J Gastroenterol. 27:4160–4171. 2021. View Article : Google Scholar | |
|
Rizvi A, Patel Z, Liu Y, Satapathy SK, Sultan K and Trindade AJ; Northwell Health COVID-19 Research Consortium: Gastrointestinal sequelae 3 and 6 months after hospitalization for coronavirus disease 2019. Clin Gastroenterol Hepatol. 19:2438–2440.e1. 2021. View Article : Google Scholar : | |
|
Fang LG and Zhou Q: Remarkable gastrointestinal and liver manifestations of COVID-19: A clinical and radiologic overview. World J Clin Cases. 9:4969–4979. 2021. View Article : Google Scholar : | |
|
Gkogkou E, Barnasas G, Vougas K and Trougakos IP: Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 36:1016152020. View Article : Google Scholar | |
|
Jackson CB, Farzan M, Chen B and Choe H: Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 23:3–20. 2022. View Article : Google Scholar | |
|
Parmar MS: TMPRSS2: An equally important protease as ACE2 in the pathogenicity of SARS-CoV-2 Infection. Mayo Clin Proc. 96:2748–2752. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Huang X, He C, Hua X, Kan A, Sun S, Wang J and Li S: Bioinformatic Analysis of correlation between immune infiltration and COVID-19 in cancer patients. Int J Biol Sci. 16:2464–2476. 2020. View Article : Google Scholar : | |
|
Hoang T, Nguyen TQ and Tran TTA: Genetic Susceptibility of ACE2 and TMPRSS2 in six common cancers and possible impacts on COVID-19. Cancer Res Treat. 53:650–656. 2021. View Article : Google Scholar | |
|
Viveiros A, Gheblawi M, Aujla PK, Sosnowski DK, Seubert JM, Kassiri Z and Oudit GY: Sex- and age-specific regulation of ACE2: Insights into severe COVID-19 susceptibility. J Mol Cell Cardiol. 164:13–16. 2022. View Article : Google Scholar | |
|
Da Eira D, Jani S and Ceddia RB: Obesogenic and ketogenic diets distinctly regulate the SARS-CoV-2 Entry Proteins ACE2 and TMPRSS2 and the Renin-angiotensin system in rat lung and heart tissues. Nutrients. 13:33572021. View Article : Google Scholar | |
|
Rando HM, MacLean AL, Lee AJ, Lordan R, Ray S, Bansal V, Skelly AN, Sell E, Dziak JJ, Shinholster L, et al: Pathogenesis, symptomatology, and transmission of SARS-CoV-2 through analysis of viral genomics and structure. mSystems. 6:e00095212021. View Article : Google Scholar | |
|
Saied EM, El-Maradny YA, Osman AA, Darwish AMG, Abo Nahas HH, Niedbala G, Piekutowska M, Abdel-Rahman MA, Balbool BA and Abdel-Azeem AM: A comprehensive review about the molecular structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Insights into natural products against COVID-19. Pharmaceutics. 13:17592021. View Article : Google Scholar : | |
|
Salem R, El-Kholy AA, Waly FR, Ayman D, Sakr A and Hussein M: Generation and utility of a single-chain fragment variable monoclonal antibody platform against a baculovirus expressed recombinant receptor binding domain of SARS-CoV-2 spike protein. Mol Immunol. 141:287–296. 2022. View Article : Google Scholar | |
|
Tai L, Zhu G, Yang M, Cao L, Xing X, Yin G, Chan C, Qin C, Rao Z, Wang X, et al: Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein. Proc Natl Acad Sci USA. 118:e21127031182021. View Article : Google Scholar : | |
|
Grishin AM, Dolgova NV, Landreth S, Fisette O, Pickering IJ, George GN, Falzarano D and Cygler M: Disulfide bonds play a critical role in the structure and function of the receptor-binding domain of the SARS-CoV-2 spike antigen. J Mol Biol. 434:1673572022. View Article : Google Scholar | |
|
Chen Y, Guo Y, Pan Y and Zhao ZJ: Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 525:135–140. 2020. View Article : Google Scholar : | |
|
Zhang J, Xiao T, Cai Y and Chen B: Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 50:173–182. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Edenfield RC and Easley CA IV: Implications of testicular ACE2 and the renin-angiotensin system for SARS-CoV-2 on testis function. Nat Rev Urol. 19:116–127. 2022. View Article : Google Scholar | |
|
Li D, Liu X, Zhang L, He J, Chen X, Liu S, Fu J, Fu S, Chen H, Fu J and Cheng J: COVID-19 disease and malignant cancers: The impact for the furin gene expression in susceptibility to SARS-CoV-2. Int J Biol Sci. 17:3954–3967. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, et al: The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 6:899–909. 2021. View Article : Google Scholar | |
|
Wu CT, Lidsky PV, Xiao Y, Lee IT, Cheng R, Nakayama T, Jiang S, Demeter J, Bevacqua RJ, Chang CA, et al: SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 33:1565–1576.e5. 2021. View Article : Google Scholar | |
|
Yele V, Sanapalli BKR and Mohammed AA: Imidazoles and benzimidazoles as putative inhibitors of SARS-CoV-2 B.1.1.7 (Alpha) and 1 (Gamma) variant spike glycoproteins: A computational approach. Chem Zvesti. 76:1107–1117. 2022. | |
|
Liu C, Zhou D, Nutalai R, Duyvesteyn HME, Tuekprakhon A, Ginn HM, Dejnirattisai W, Supasa P, Mentzer AJ, Wang B, et al: The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Cell Host Microbe. 30:53–68.e12. 2022. View Article : Google Scholar | |
|
Moss DL and Rappaport J: SARS-CoV-2 beta variant substitutions alter spike glycoprotein receptor binding domain structure and stability. J Biol Chem. 297:1013712021. View Article : Google Scholar : PubMed/NCBI | |
|
Sanches PRS, Charlie-Silva I, Braz HLB, Bittar C, Freitas Calmon M, Rahal P and Cilli EM: Recent advances in SARS-CoV-2 Spike protein and RBD mutations comparison between new variants Alpha (B.1.1.7, United Kingdom), Beta (B.1.351, South Africa), Gamma (1Brazil) and Delta (B.1.617.2, India). J Virus Erad. 7:1000542021. View Article : Google Scholar | |
|
Storti B, Quaranta P, Di Primio C, Clementi N, Mancini N, Criscuolo E, Spezia PG, Carnicelli V, Lottini G, Paolini E, et al: A spatial multi-scale fluorescence microscopy toolbox discloses entry checkpoints of SARS-CoV-2 variants in Vero E6 cells. Comput Struct Biotechnol J. 19:6140–6156. 2021. View Article : Google Scholar : | |
|
Alaofi AL and Shahid M: Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules. 11:12732021. View Article : Google Scholar : | |
|
Bhattacharya M, Chatterjee S, Sharma AR, Agoramoorthy G and Chakraborty C: D614G mutation and SARS-CoV-2: Impact on S-protein structure, function, infectivity, and immunity. Appl Microbiol Biotechnol. 105:9035–9045. 2021. View Article : Google Scholar : | |
|
Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, et al: First Case of 2019 novel coronavirus in the United States. N Engl J Med. 382:929–936. 2020. View Article : Google Scholar : | |
|
Wang W, Xu Y, Gao R, Lu R, Han K, Wu G and Tan W: Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 323:1843–1844. 2020. | |
|
Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, et al: Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 26:502–505. 2020. View Article : Google Scholar | |
|
Chen L, Lou J, Bai Y and Wang M: COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. Am J Gastroenterol. 115:7902020. View Article : Google Scholar : PubMed/NCBI | |
|
Burgueño JF, Reich A, Hazime H, Quintero MA, Fernandez I, Fritsch J, Santander AM, Brito N, Damas OM, Deshpande A, et al: Expression of SARS-CoV-2 Entry Molecules ACE2 and TMPRSS2 in the Gut of Patients With IBD. Inflamm Bowel Dis. 26:797–808. 2020. View Article : Google Scholar | |
|
Xiao F, Tang M, Zheng X, Liu Y, Li X and Shan H: Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 158:1831–1833.e3. 2020. View Article : Google Scholar | |
|
Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, et al: Extrapulmonary manifestations of COVID-19. Nat Med. 26:1017–1032. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Mohamed DZ, Ghoneim ME, Abu-Risha SE, Abdelsalam RA and Farag MA: Gastrointestinal and hepatic diseases during the COVID-19 pandemic: Manifestations, mechanism and management. World J Gastroenterol. 27:4504–4535. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, Gu Z, Gao L, Shi H, Mai L, et al: Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 69:997–1001. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Elmunzer BJ, Spitzer RL, Foster LD, Merchant AA, Howard EF, Patel VA, West MK, Qayed E, Nustas R, Zakaria A, et al: Digestive manifestations in patients hospitalized with coronavirus disease 2019. Clin Gastroenterol Hepatol. 19:1355–1365.e4. 2021. View Article : Google Scholar | |
|
Ferm S, Fisher C, Pakala T, Tong M, Shah D, Schwarzbaum D, Cooley V, Hussain S and Kim SH: Analysis of gastrointestinal and hepatic manifestations of SARS-CoV-2 infection in 892 patients in queens, NY. Clin Gastroenterol Hepatol. 18:2378–2379.e1. 2020. View Article : Google Scholar | |
|
Wang MK, Yue HY, Cai J, Zhai YJ, Peng JH, Hui JF, Hou DY, Li WP and Yang JS: COVID-19 and the digestive system: A comprehensive review. World J Clin Cases. 9:3796–3813. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Andrews PLR, Cai W, Rudd JA and Sanger GJ: COVID-19, nausea, and vomiting. J Gastroenterol Hepatol. 36:646–656. 2021. View Article : Google Scholar | |
|
Boraschi P, Giugliano L, Mercogliano G, Donati F, Romano S and Neri E: Abdominal and gastrointestinal manifestations in COVID-19 patients: Is imaging useful? World J Gastroenterol. 27:4143–4159. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Carvalho A, Alqusairi R, Adams A, Paul M, Kothari N, Peters S and DeBenedet AT: SARS-CoV-2 gastrointestinal infection causing hemorrhagic colitis: Implications for detection and transmission of COVID-19 disease. Am J Gastroenterol. 115:942–946. 2020. View Article : Google Scholar | |
|
Li X, Huang S, Lu J, Lai R, Zhang Z, Lin X, Zheng X and Shan H: Upper Gastrointestinal Bleeding Caused by SARS-CoV-2 Infection. Am J Gastroenterol. 115:1541–1542. 2020. View Article : Google Scholar | |
|
Xu Z, Tang M, Chen P, Cai H and Xiao F: SARS-CoV-2 gastro-intestinal infection prolongs the time to recover from COVID-19. Front Med (Lausanne). 8:6835512021. View Article : Google Scholar | |
|
Hu F, Chen F, Ou Z, Fan Q, Tan X, Wang Y, Pan Y, Ke B, Li L, Guan Y, et al: A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract. Cell Mol Immunol. 17:1119–1125. 2020. View Article : Google Scholar | |
|
Noviello D, Costantino A, Muscatello A, Bandera A, Consonni D, Vecchi M and Basilisco G: Functional gastrointestinal and somatoform symptoms five months after SARS-CoV-2 infection: A controlled cohort study. Neurogastroenterol Motil. 34:e141872022. View Article : Google Scholar | |
|
Liu YL, Ren J, Yuan JP, Zhang ZJ, Guo WY, Guan Y, Moeckel G, Ahuja N and Fu T: Postoperative onset and detection of SARS-CoV-2 in surgically resected specimens from gastrointestinal cancer patients with pre/asymptomatic COVID-19. Ann Surg. 272:e321–e328. 2020. View Article : Google Scholar | |
|
Nabil A, Elshemy MM, Uto K, Soliman R, Hassan AA, Shiha G and Ebara M: Coronavirus (SARS-CoV-2) in gastroenterology and its current epidemiological situation: An updated review until January 2021. EXCLI J. 20:366–385. 2021.PubMed/NCBI | |
|
McAllister MJ, Kirkwood K, Chuah SC, Thompson EJ, Cartwright JA, Russell CD, Dorward DA, Lucas CD and Ho GT: Intestinal protein characterisation of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in inflammatory bowel disease (IBD) and Fatal COVID-19 Infection. Inflammation. 45:567–572. 2022. View Article : Google Scholar | |
|
Suárez-Fariñas M, Tokuyama M, Wei G, Huang R, Livanos A, Jha D, Levescot A, Irizar H, Kosoy R, Cording S, et al: Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2-related disease. Gastroenterology. 160:287–301.e20. 2021. View Article : Google Scholar | |
|
Tao SS, Wang XY, Yang XK, Liu YC, Fu ZY, Zhang LZ, Wang ZX, Ni J, Shuai ZW and Pan HF: COVID-19 and inflammatory bowel disease crosstalk: From emerging association to clinical proposal. J Med Virol. 94:5640–5652. 2022. View Article : Google Scholar | |
|
Shen S, Gong M, Wang G, Dua K, Xu J, Xu X and Liu G: COVID-19 and gut injury. Nutrients. 14:44092022. View Article : Google Scholar | |
|
Viganò C, Massironi S, Pirola L, Cristoferi L, Fichera M, Bravo M, Mauri M, Redaelli AE, Dinelli ME and Invernizzi P: COVID-19 in patients with inflammatory bowel disease: A single-center observational study in Northern Italy. Inflamm Bowel Dis. 26:e138–e139. 2020. View Article : Google Scholar | |
|
Derikx LAAP, Lantinga MA, de Jong DJ, van Dop WA, Creemers RH, Römkens TEH, Jansen JM, Mahmmod N, West RL, Tan ACITL, et al: Clinical Outcomes of Covid-19 in patients with inflammatory bowel disease: A nationwide cohort study. J Crohns Colitis. 15:529–539. 2021. View Article : Google Scholar | |
|
Zhou L, Niu Z, Jiang X, Zhang Z, Zheng Y, Wang Z, Zhu Y, Gao L, Huang H, Wang X and Sun Q: SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. iScience. 23:1017442020. View Article : Google Scholar : PubMed/NCBI | |
|
Qi F, Qian S, Zhang S and Zhang Z: Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 526:135–140. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Lee JJ, Kopetz S, Vilar E, Shen JP, Chen K and Maitra A: Relative Abundance of SARS-CoV-2 entry genes in the enterocytes of the lower gastrointestinal tract. Genes (Basel). 11:6452020. View Article : Google Scholar : PubMed/NCBI | |
|
An X, Lin W, Liu H, Zhong W, Zhang X, Zhu Y, Wang X, Li J and Sheng Q: SARS-CoV-2 Host Receptor ACE2 protein expression atlas in human gastrointestinal tract. Front Cell Dev Biol. 9:6598092021. View Article : Google Scholar | |
|
Zhang M, Feng C, Zhang X, Hu S, Zhang Y, Min M, Liu B, Ying X and Liu Y: Susceptibility factors of stomach for SARS-CoV-2 and treatment implication of mucosal protective agent in COVID-19. Front Med (Lausanne). 7:5979672021. View Article : Google Scholar : PubMed/NCBI | |
|
Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, et al: A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 28:124–133.e4. 2020. View Article : Google Scholar | |
|
Hartman AL, Nambulli S, McMillen CM, White AG, Tilston-Lunel NL, Albe JR, Cottle E, Dunn MD, Frye LJ, Gilliland TH, et al: SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog. 16:e10089032020. View Article : Google Scholar : PubMed/NCBI | |
|
Jiao L, Li H, Xu J, Yang M, Ma C, Li J, Zhao S, Wang H, Yang Y, Yu W, et al: The gastrointestinal tract is an alternative route for SARS-CoV-2 Infection in a nonhuman primate model. Gastroenterology. 160:1647–1661. 2021. View Article : Google Scholar | |
|
Livanos AE, Jha D, Cossarini F, Gonzalez-Reiche AS, Tokuyama M, Aydillo T, Parigi TL, Ladinsky MS, Ramos I, Dunleavy K, et al: Intestinal host response to SARS-CoV-2 Infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 160:2435–2450.e34. 2021. View Article : Google Scholar | |
|
Qi J, Zhou Y, Hua J, Zhang L, Bian J, Liu B, Zhao Z and Jin S: The scRNA-seq Expression Profiling of the Receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to SARS-CoV-2 Infection. Int J Environ Res Public Health. 18:2842021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang M, Yan W, Qi W, Wu D, Zhu L, Li W, Wang X, Ma K, Ni M, Xu D, et al: Clinical characteristics and risk factors of liver injury in COVID-19: A retrospective cohort study from Wuhan, China. Hepatol Int. 14:723–732. 2020. View Article : Google Scholar | |
|
Zhang H, Liao YS, Gong J, Liu J and Zhang H: Clinical characteristics and risk factors for liver injury in COVID-19 patients in Wuhan. World J Gastroenterol. 26:4694–4702. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wisniewska H, Skowron M, Bander D, Hornung M, Jurczyk K, Karpinska E, Laurans Ł, Socha Ł, Czajkowski Z and Wawrzynowicz-Syczewska M: Nosocomial COVID-19 Infection and Severe COVID-19 pneumonia in patients hospitalized for alcoholic liver disease: A case report. Am J Case Rep. 21:e9274522020. View Article : Google Scholar : PubMed/NCBI | |
|
Yang SJ, Wei TC, Hsu CH, Ho SN, Lai CY, Huang SF, Chen YY, Liu SJ, Yu GY and Dou HY: Characterization of virus replication, pathogenesis, and cytokine responses in syrian hamsters inoculated with SARS-CoV-2. J Inflamm Res. 14:3781–3795. 2021. View Article : Google Scholar | |
|
Wong GL, Yip TC, Wong VW, Tse YK, Hui DS, Lee SS, Yeoh EK, Chan HL and Lui GC: SARS-CoV-2 viral persistence based on cycle threshold value and liver injury in patients with COVID-19. Open Forum Infect Dis. 8:ofab2052021. View Article : Google Scholar | |
|
Lei HY, Ding YH, Nie K, Dong YM, Xu JH, Yang ML, Liu MQ, Wei L, Nasser MI, Xu LY, et al: Potential effects of SARS-CoV-2 on the gastrointestinal tract and liver. Biomed Pharmacother. 133:1110642021. View Article : Google Scholar | |
|
Zhong P, Xu J, Yang D, Shen Y, Wang L, Feng Y, Du C, Song Y, Wu C, Hu X and Sun Y: COVID-19-associated gastrointestinal and liver injury: Clinical features and potential mechanisms. Signal Transduct Target Ther. 5:2562020. View Article : Google Scholar : | |
|
Siddiqui MA, Suresh S, Simmer S, Abu-Ghanimeh M, Karrick M, Nimri F, Musleh M, Mediratta V, Al-Shammari M, Russell S, et al: Increased morbidity and mortality in COVID-19 patients with liver injury. Dig Dis Sci. 67:2577–2583. 2021. View Article : Google Scholar | |
|
Wijarnpreecha K, Ungprasert P, Panjawatanan P, Harnois DM, Zaver HB, Ahmed A and Kim D: COVID-19 and liver injury: A meta-analysis. Eur J Gastroenterol Hepatol. 33:990–995. 2021. View Article : Google Scholar | |
|
Wang Q, Zhao H, Liu LG, Wang YB, Zhang T, Li MH, Xu YL, Gao GJ, Xiong HF, Fan Y, et al: Pattern of liver injury in adult patients with COVID-19: A retrospective analysis of 105 patients. Mil Med Res. 7:282020.PubMed/NCBI | |
|
Del Nonno F, Nardacci R, Colombo D, Visco-Comandini U, Cicalini S, Antinori A, Marchioni L, D'Offizi G, Piacentini M and Falasca L: Hepatic failure in COVID-19: Is iron overload the dangerous trigger? Cells. 10:11032021. View Article : Google Scholar : PubMed/NCBI | |
|
Gomi K, Ito T, Yamaguchi F, Kamio Y, Sato Y, Mori H, Endo K, Abe T, Sakakura S, Kobayashi K, et al: Clinical features and mechanism of liver injury in patients with mild or moderate coronavirus disease 2019. JGH Open. 5:888–895. 2021. View Article : Google Scholar | |
|
Ma C, Cong Y and Zhang H: COVID-19 and the digestive system. Am J Gastroenterol. 115:1003–1006. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Q, Li J, Zhang Y, Gao J, Wang P, Ai M, Ding W and Tan X: Differences in clinical characteristics and liver injury between suspected and confirmed COVID-19 patients in Jingzhou, Hubei Province of China. Medicine (Baltimore). 100:e259132021. View Article : Google Scholar : PubMed/NCBI | |
|
Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, et al: Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 383:120–128. 2020. View Article : Google Scholar : | |
|
Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F and Moch H: Endothelial cell infection and endotheliitis in COVID-19. Lancet. 395:1417–1418. 2020. View Article : Google Scholar : | |
|
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al: Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 8:420–422. 2020. View Article : Google Scholar : | |
|
Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM and Angus PW: Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 54:1790–1796. 2005. View Article : Google Scholar | |
|
Bender JM and Worman HJ: Coronavirus Disease 2019 and liver injury: A retrospective analysis of hospitalized patients in New York City. J Clin Transl Hepatol. 9:551–558. 2021.PubMed/NCBI | |
|
Chew M, Tang Z, Radcliffe C, Caruana D, Doilicho N, Ciarleglio MM, Deng Y and Garcia-Tsao G: Significant liver injury during hospitalization for COVID-19 is not associated with liver insufficiency or death. Clin Gastroenterol Hepatol. 19:2182–2191.e7. 2021. View Article : Google Scholar | |
|
Vishwajeet V, Purohit A, Kumar D, Parag V, Tripathi S, Kanchan T, Kothari N, Dutt N, Elhence PA, Bhatia PK, et al: Evaluation of pathological findings of COVID-19 by minimally invasive autopsies: A single tertiary care center experience from India. J Lab Physicians. 13:97–106. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Tollard C, Champenois V, Delemer B, Carsin-Vu A and Barraud S: An inaugural diabetic ketoacidosis with acute pancreatitis during COVID-19. Acta Diabetol. 58:389–391. 2021. View Article : Google Scholar | |
|
Kumaran NK, Karmakar BK and Taylor OM: Coronavirus disease-19 (COVID-19) associated with acute necrotising pancreatitis (ANP). BMJ Case Rep. 13:e2379032020. View Article : Google Scholar : PubMed/NCBI | |
|
Alves AM, Yvamoto EY, Marzinotto MAN, Teixeira ACS and Carrilho FJ: SARS-CoV-2 leading to acute pancreatitis: An unusual presentation. Braz J Infect Dis. 24:561–564. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Hadi A, Werge M, Kristiansen KT, Pedersen UG, Karstensen JG, Novovic S and Gluud LL: Coronavirus Disease-19 (COVID-19) associated with severe acute pancreatitis: Case report on three family members. Pancreatology. 20:665–667. 2020. View Article : Google Scholar | |
|
Barlass U, Wiliams B, Dhana K, Adnan D, Khan SR, Mahdavinia M and Bishehsari F: Marked elevation of lipase in COVID-19 Disease: A cohort study. Clin Transl Gastroenterol. 11:e002152020. View Article : Google Scholar | |
|
Inamdar S, Benias PC, Liu Y, Sejpal DV, Satapathy SK and Trindade AJ; Northwell COVID-19 Research Consortium: Prevalence, risk factors, and outcomes of hospitalized patients with coronavirus disease 2019 presenting as acute pancreatitis. Gastroenterology. 159:2226–2228.e2. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Samies NL, Yarbrough A and Boppana S: Pancreatitis in pediatric patients with COVID-19. J Pediatric Infect Dis Soc. 10:57–59. 2021. View Article : Google Scholar | |
|
Pandanaboyana S, Moir J, Leeds JS, Oppong K, Kanwar A, Marzouk A, Belgaumkar A, Gupta A, Siriwardena AK, Haque AR, et al: SARS-CoV-2 infection in acute pancreatitis increases disease severity and 30-day mortality: COVID PAN collaborative study. Gut. 70:1061–1069. 2021. View Article : Google Scholar | |
|
Liu F, Long X, Zhang B, Zhang W, Chen X and Zhang Z: ACE2 expression in pancreas may cause pancreatic damage after SARS-CoV-2 Infection. Clin Gastroenterol Hepatol. 18:2128–2130.e2. 2020. View Article : Google Scholar : | |
|
Qadir MMF, Bhondeley M, Beatty W, Gaupp DD, Doyle-Meyers LA, Fischer T, Bandyopadhyay I, Blair RV, Bohm R, Rappaport J, et al: SARS-CoV-2 infection of the pancreas promotes thrombofibrosis and is associated with new-onset diabetes. JCI Insight. 6:e1515512021. View Article : Google Scholar : | |
|
Jablonska B, Olakowski M and Mrowiec S: Association between acute pancreatitis and COVID-19 infection: What do we know? World J Gastrointest Surg. 13:548–562. 2021. View Article : Google Scholar : | |
|
Cao W, Feng Q and Wang X: Computational analysis of TMPRSS2 expression in normal and SARS-CoV-2-infected human tissues. Chem Biol Interact. 346:1095832021. View Article : Google Scholar : | |
|
Kusmartseva I, Wu W, Syed F, Van Der Heide V, Jorgensen M, Joseph P, Tang X, Candelario-Jalil E, Yang C, Nick H, et al: Expression of SARS-CoV-2 entry factors in the pancreas of normal organ donors and individuals with COVID-19. Cell Metab. 32:1041–1051.e6. 2020. View Article : Google Scholar : | |
|
Coate KC, Cha J, Shrestha S, Wang W, Goncalves LM, Almaca J, Kapp ME, Fasolino M, Morgan A, Dai C, et al: SARS-CoV-2 cell entry factors ACE2 and TMPRSS2 are expressed in the microvasculature and ducts of human pancreas but are not enriched in β cells. Cell Metab. 32:1028–1040.e4. 2020. View Article : Google Scholar | |
|
Steenblock C, Richter S, Berger I, Barovic M, Schmid J, Schubert U, Jarzebska N, von Mässenhausen A, Linkermann A, Schürmann A, et al: Viral infiltration of pancreatic islets in patients with COVID-19. Nat Commun. 12:35342021. View Article : Google Scholar | |
|
Shaharuddin SH, Wang V, Santos RS, Gross A, Wang Y, Jawanda H, Zhang Y, Hasan W, Garcia G Jr, Arumugaswami V and Sareen D: Deleterious Effects of SARS-CoV-2 infection on human pancreatic cells. Front Cell Infect Microbiol. 11:6784822021. View Article : Google Scholar |