Single‑cell sequencing, genetics, and epigenetics reveal mesenchymal stem cell senescence in osteoarthritis (Review)
- Authors:
- Published online on: November 7, 2023 https://doi.org/10.3892/ijmm.2023.5326
- Article Number: 2
-
Copyright: © Tan et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
1. Introduction
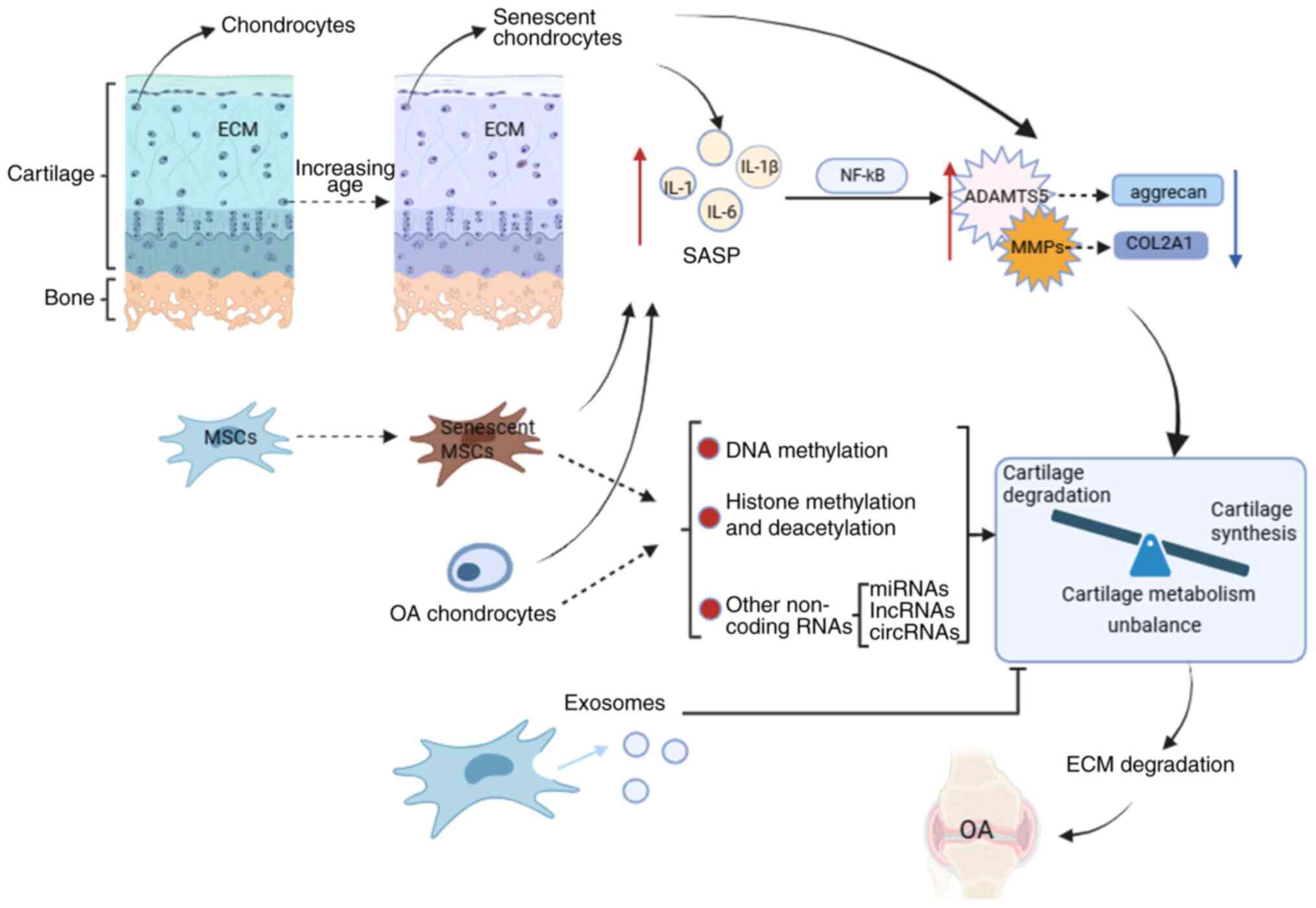
Osteoarthritis (OA) is a chronic joint disease characterized by articular cartilage degeneration and secondary bone hyperplasia. The disease affects the articular cartilage or the entire joint, including the subchondral bone, joint capsule, synovium and muscles around the joint (1). The cause of primary OA remains not fully understood, and its occurrence and development is a long-term, chronic and progressive pathological process. OA is often considered to be the result of an interaction of multiple pathogenic factors, including mechanical and biological factors. Among them, age is the main risk factor, and the other factors include trauma, obesity, genetics, inflammation and metabolism (2). The earliest and most important pathological changes in OA occur in cartilage. Mesenchymal stem cells (MSCs) can differentiate into chondrocytes (3). Therefore, MSC aging may be related to the occurrence and progression of OA. The present review aimed to describe the association between OA and cellular aging in terms of genetics, epigenetics, and single-cell sequencing, focusing on the association between the aging of MSC and OA (Fig. 1).
2. Cellular senescence
The main types of aging include replication, DNA damage-induced, oncogene-induced, oxidative stress-induced, chemotherapy-induced, mitochondrial dysfunction-associated, epigenetically-induced [DNA methylase inhibitors or histone deacetylase (HDAC) are also known to cause senescence], and paracrine senescence (senescence-associated secretory phenotype-induced senescence) produced by primary senescent cells (4). Cellular senescence refers to the permanent growth arrest in cells with the ability to proliferate and respond to various cellular stresses. It promotes tissue remodeling during development and after an injury, but may also lead to decreased regenerative potential and function of tissues and tumorigenesis of inflammatory and senescent organisms (4). Markers of aging include genomic instability, telomere depletion, epigenetic alterations, loss of protein balance, malfunction of nutrient perception, mitochondrial dysfunction, cellular senescence, stem cell failure and altered intercellular communication (5). Numerous factors associated with aging, such as mitochondrial dysfunction, oxidative stress and cellular senescence, contribute to OA.
Cellular senescence is one of the hallmarks of senescence, and a key feature of senescent cells is the secretion of a range of pro-inflammatory cytokines, chemokines and growth factors such as interleukin (IL)-6, IL-1β, known as the senescence-associated secretory phenotype (SASP) (6). SASP is observed in numerous senescent cell types, including fibroblasts and MSCs (7,8). Phenotypic genes (IL-1β, IL-6, IL-7, IL-8, MMP family) associated with senescent SASP show increased expression in cartilage and synovial tissue in patients with OA (9). These SASP factors may be produced in part by OA chondrocytes (10). In animal experiments, expression levels of the inflammatory markers matrix metalloproteinase 13 (MMP13) and IL-1β were reduced when senescent cells were removed by transgenic mouse models or drug intervention (11). Both expressions are manifestations of OA. Therefore, OA may be associated with the senescence of chondrocytes.
Stem cells can be roughly divided into embryonic stem cells and adult stem cells according to the source, and MSCs belong to adult stem cells, which are stromal cells with a self-renewal ability and exhibit multidirectional differentiation. MSCs can be isolated from a variety of tissues, including the umbilical cord, endometrial polyps, menstrual blood, bone marrow and adipose tissue (12). MSCs, with a multidirectional differentiation capacity, can differentiate into all mesoderm-derived cell types, such as fat cells, osteoblasts and chondrocytes (3). The function of MSCs declines with age, a process known as aging. This may be associated with a loss of tissue homeostasis maintenance, leading to organ failure and aging diseases (13,14) such as OA. The clinical value of MSCs comes primarily from their non-stem cell/progenitor cell properties. That is, MSCs produce extracellular vesicles, including exosomes (described below), several cytokines and growth factors involved in regulating tissue metabolism and tissue repair and regeneration after injury (15), as in OA.
3. Molecular landscape of OA
OA affects joint cartilage or the entire joint, and cartilage damage is an important part of the development of OA. Potential mechanisms that contribute to OA include age-related inflammation, cellular senescence (including SASP), mitochondrial dysfunction and oxidative stress, energy metabolism dysfunction (associated with decreased autophagy) and alterations in cell signaling (16). In recent years, numerous studies have suggested that chloride ion channels may also be one of the mechanisms that lead to the development of OA (17). Cartilage damage, meaning loss of the extracellular matrix (ECM) of chondrocytes, is a key early feature of OA, characterized by inadequate ECM synthesis and degeneration of articular cartilage. Cartilage consists of a single cell type, the chondrocytes (18), which are surrounded by a large ECM. The ECM consists of two main components, mainly collagen (types II, IX and XI) and proteoglycans (mainly aggrecan) (19-21), the gene expression of which is controlled by transcription factors SRY-related HMG-box (SOX)-5, SOX-6, and SOX-9 (22). Aggrecan, a major proteoglycan in the articular cartilage, is cleaved at a specific 'aggrecanase' site in human osteoarthritic cartilage; this cleavage is performed by several members of a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family of metalloproteases, with ADAMTS-5 playing a major role (23). Matrix metalloproteinases (MMPs) are responsible for the breakdown of chondro-collagen, with MMP-13 playing a central role (24).
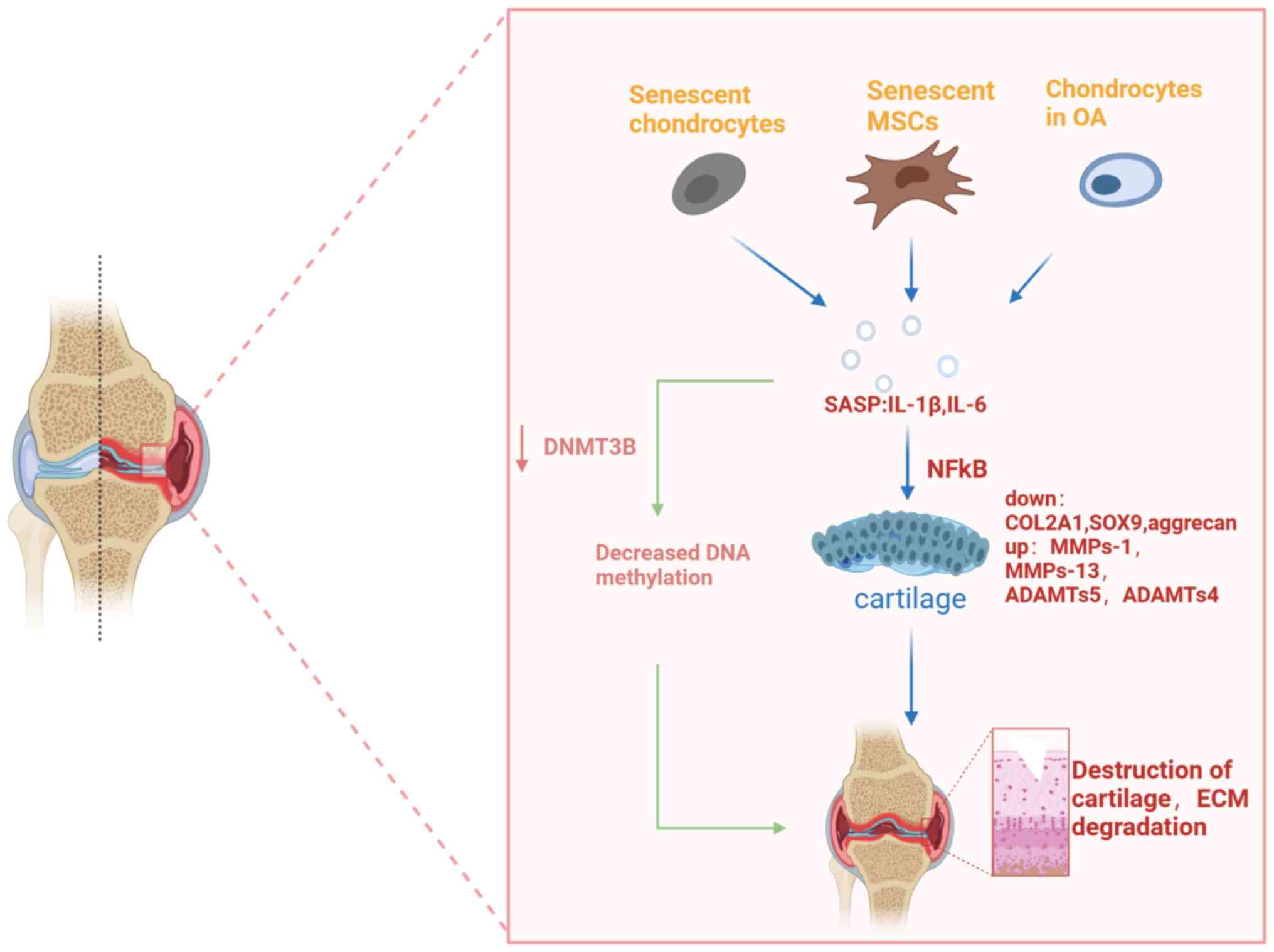
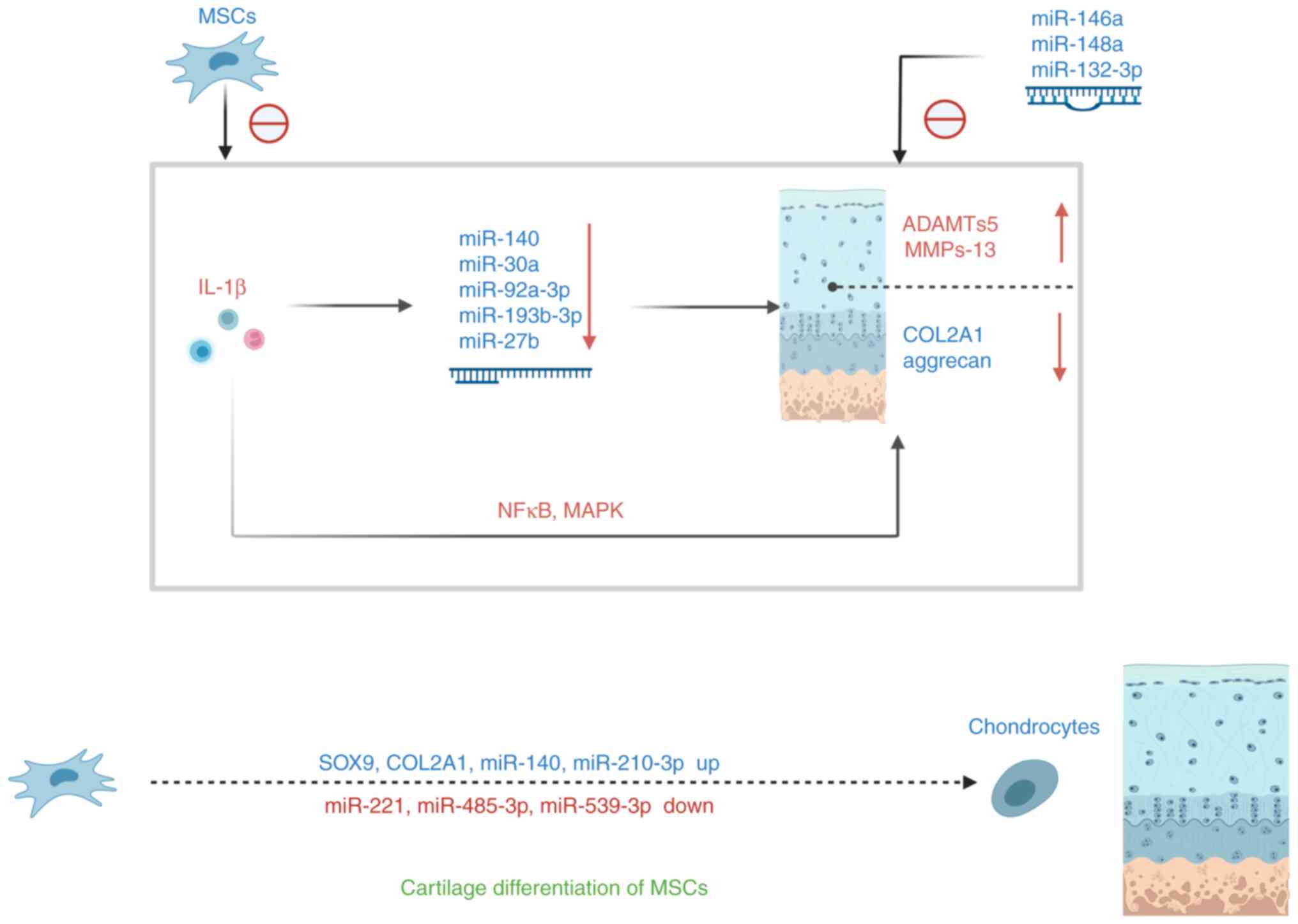
OA is characterized by accumulation of senescent cells and the wearing off of the protective cartilage. A previous study showed an increase in the number of senescent cells in articular cartilage, synovial membrane, and fat pad tissues during OA progression (25). The development and progression of OA may be caused by the accumulation of senescent cells within or near the joint (6). With age, the proliferation and synthesis capacity of articular chondrocytes decreases, the aging chondrocytes are unable to produce and repair the ECM (26). Characteristic changes in senescent chondrocytes include increased production of cytokines (for example, IL-1β, IL-1 and IL-6) and MMP, increased levels of senescence-associated beta-galactosidase, p53, p21 and p16 and decreased collagen II (COL2) synthesis and sirtuin 1 (SIRT1) (10,26). In OA, joint cartilage degradation triggers an inflammatory response and cytokine production in the tissues around the joint. These inflammatory molecules stimulate further ECM catabolism by increasing protease synthesis in chondrocytes (27). Senescent fibroblasts and MSCs can also produce a range of pro-inflammatory cytokines (7,8). In addition, studies have shown that a decrease in the level of autophagy during aging also increases the release of pro-inflammatory factors (28). Cartilage destruction begins when chondrocytes are stimulated by pro-inflammatory cytokines such as IL-1β, IL-6, IL-8 and tumor necrosis factor (TNF)-α (29,30). Disruption of cartilage stromal integrity is caused by increased catabolism/apoptosis of chondrocytes in articular cartilage and decreased chondrocyte anabolism (31,32). IL-1β and TNF-α are among the key pro-inflammatory cytokines involved cartilage destruction in OA (27). Numerous studies have shown that elevated levels of IL-1β can be observed in the synovial fluid, synovium, subchondral bone and cartilage in patients with OA (33). Numerous studies have confirmed that IL-1β interferes with the synthesis of key structural proteins such as type II collagen and aggrecan (34,35). IL-1β induced enzymes of the MMP family, mainly interstitial collagenase (MMP-1), streptoglobulinase-1 (MMP-3), and collagenase 3 (MMP-13). These enzymes affect the expression of cartilage collagen (24,36), thereby affecting the balance between cartilage matrix synthesis and catabolism, leading to degenerative cartilage diseases such as OA (32). In addition, IL-1β induces chondrocytes to produce ADAMTS metalloproteinases (36), which affects aggrecan expression and regulates cartilage metabolism (37). A previous study suggested that IL-1β-induced chloride channel opening may also be strongly associated with the development of OA (38). The pro-inflammatory and catabolic effects of IL-1β and TNF are mediated by the activation of several signaling pathways, most importantly, the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) signaling pathway (39). NFκB mediates the expression of several inflammatory genes, such as those encoding inducible nitric oxide synthase (iNOS), cyclooxygenase 2 (COX2) and chemokines, and also helps induce MMP-1, MMP-9, MMP-13 and ADAMTS4 (39,40). IL-1β significantly upregulated the expression of MMP-13, NFκB, p65, etc., inhibiting the expression of COL2 and aggrecan (ACAN) (41). In contrast, MSCs enhanced the expression of COL2 and aggrecan and inhibited the expression of MMP-13 and NFκB p65 in IL-1β-stimulated rat chondrocytes, partially through the NFκB signaling pathway (41) (Fig. 2).
Additionally, elevated levels of TNF-α, IL-1, and IL-6 in synovial fluid, synovium, subchondral bone, and cartilage in patients with OA induce the production of other cytokines, MMPs, and prostaglandins, and inhibit the synthesis of proteoglycans and COL2. Chondrocytes activated by these pro-inflammatory cytokines in turn produce MMP-1, MMP-3, MMP-13, and aggrecanase 1 and 2 (ADAMTS-4 and ADAMTS-5, respectively) expression (19). This in turn causes cartilage degradation. Senescent cells also produce SASPs such as IL-1β, IL-6 and TNF-α, which are key pro-inflammatory cytokines associated with OA (19). Therefore, they play a key role in cartilage matrix degradation and bone resorption in OA (42). The increase in inflammatory factors above in OA reflects the senescence of chondrocytes (characteristic changes in the senescence of chondrocytes). Similarly, aging MSCs also produce SASP, which affects cartilage-specific gene expression and inhibits cartilage catabolic gene expression, thereby promoting cartilage breakdown and reducing cartilage synthesis, which is related to the occurrence and development of OA.
The transcription factor SOX9 is essential for chondrocyte differentiation and cartilage formation (43,44). Sox9 was the main positive regulator of chondrogenesis, and its regulation affected cartilage differentiation by MSCs in a mouse model (45,46). SOX9 controlled the expression of numerous chondrocyte genes, including its cofactors L-SOX5a and SOX6, as well as ECM genes, such as type II collagen and aggrecan (47), involved in regulating cartilage formation. Chondrogenic RNA modulator (ROCR), a long non-coding RNA (lncRNA) necessary for successful differentiation of MSCs into chondrocytes, appeared to aid in the expression of SOX9. Normalizing SOX9 levels by overexpression reversed the damaged cartilage phenotype caused by ROCR depletion (47). SOX9, on the other hand, promoted microRNA (miR)-140 expression (48,49), which in turn promotes collagen II expression and inhibits MMP-13 and ADAMTS-5 expression (50), thereby blocking OA. However, studies have shown no significant correlation between SOX9 expression in bone marrow and cellular senescence. Association between aging MSCs and SOX9 expression needs further study (51). Nuclear factor kappa-B ligands (RANKL) are produced by osteoblasts, stromal cells, T cells and other sources and activate RANK on the surface of osteoclasts and osteoclast precursors. RANK activation leads to the recruitment of the adaptor protein TRAF 6 (TNF receptor correlated factor 6), resulting in NFκB activation (52). This may cause an imbalance in cartilage metabolism. Studies have shown that RANKL mRNA increases with age in bone marrow cells, and its expression in OA and rheumatoid arthritis (RA) bone marrow cells is markedly higher than normal (51). In animal trials, transplantation of MSCs can reduce the expression of RANKL in rats by downregulating the level of IL-22, thereby improving the degree of RA bone destruction (53). The expression of RANKL is significantly increased in senescent MSCs (54). Therefore, senescent MSCs may affect OA by regulating RANKL expression. Studies have found that the expression of Runt-associated transcription factor 2 (RUNX2) is elevated in human OA cartilage (55). Runx2 is the most effective inducer of osteoblast differentiation and a major transcription factor for chondrocytes hypertrophy. Osteocalcin, Runx2 from subchondral bone accumulates in the weight-bearing area of the OA joint. This indicates an upregulation of osteoblast and pre-osteoblastic activity, possibly due to an increased need for bone formation due to excessive mechanical load and osteocyte death (56), thus promoting cartilage breakdown and aggravating OA. Studies have shown that MSCs undergo aging and spontaneous osteogenic differentiation after regular culture amplification, and Runx2 is upregulated (57). Upregulated RUNX2 may help increase the expression of its downstream catabolic target MMP-13 affecting cartilage catabolism (55). miR-105 bound to and targeted RUNX2 in chondrocytes, and the expression of miR-105 in human OA cartilage was downregulated, which was inversely correlated with the expression of RUNX2, ADAMTS7 and ADAMTS12 (58). The expression of these genes is upregulated, causing cartilage destruction. Insufficiency of ECM synthesis and articular cartilage degeneration in patients with OA, as well as downregulation of miR-140, may be associated with a decrease in SOX9.
In summary, OA chondrocytes produce a large amount of SASP, which eventually causes cartilage metabolism imbalance through different signaling pathways, that is, cartilage catabolism is greater than cartilage anabolism, resulting in cartilage destruction, which in turn aggravates OA Senescent MSCs and chondrocytes can also upregulate the expression of cartilage catabolic genes by producing SASPs, affecting cartilage metabolism (Fig. 1).
4. Genetics reveals the mechanism underlying the effect of aging MSCs on OA
Over the past decade, OA genetics has changed through the application of large-scale genome-wide association scans (GWAS). So far, more than 100 polymorphic DNA variants have been linked to OA (59). Interpreting the results of GWAS is biologically challenging, therefore translating gene discoveries into effective therapies remains elusive for now. The OA GWAS on the UK Biobank dataset is the largest to date, surveying over 77,000 OA patients and identifying 52 new risk loci (60). For example, rs75621460 (hip and/or knee OA, single variant in the 95% credible set) is an intergenic variant located downstream of CCDC97 and TGF-β1 (60). SMAD3 (known loci: rs12901372) encodes a transcriptional regulator, plays a key role in cartilage differentiation, and regulates TGF-β1 expression (61). TGF-β1 regulates articular cartilage metabolism (as aforementioned). In addition, the TGF-β1/SMAD3 pathway regulates the expression of miR-140 in OA (62), affecting OA progression. RUNX2 (known loci: rs2064630) encodes transcription factor necessary for osteoblast differentiation and chondrocytes maturation (63), and is downregulated by TGF-β1 (64). Senescent MSCs also produce pro-inflammatory cytokines and, therefore, may have partial crosstalk with genetic locus variation.
In the future, with the application of large-scale GWAS, more risk sites will be discovered, which means that more therapeutic targets will be discovered. At that time, new treatments for OA will continue to emerge.
5. Epigenetics reveals the mechanism by which the aging MSCs affect OA
Epigenetics is defined as a heritable change in gene expression caused by external or environmental factors rather than a change in the DNA sequence (65), mainly including DNA methylation, histone modification and non-coding RNA (detailed below) (65). Changes in age-related epigenetic processes may be a potential cause of delayed human diseases such as OA. In 2017, Simon and Jeffries summarized epigenetic changes in OA such as low methylation, low HDAC-1 level, increased HDAC-2 level and downregulated miR-140 (66).
DNA methylation
DNA methylation is a reversible process catalyzed by DNA methyltransferases (DNMTs) resulting in the formation of 5-methylcytosine (5 mC), that is, methylation on the fifth carbon of the cytosine residue at the CpG dinucleotide (67). Overall, DNA methylation in OA is reduced (66). During MSC cartilage development, considerable demethylation changes occur in the epigenetic landscape, especially at sites characterized by enhancer modifications (68). Differential DNA methylation of inflammatory factors is associated with OA in human chondrocytes. For example, IL8 showed 38-fold higher expression in patients with hip OA than in the control group, and in vitro DNA methylation was noted to reduce basal IL8 promoter basal activity. Upregulation of IL-8 reduces the expression of DNMT3B in OA chondrocytes (69), causing decreased DNA methylation. The pro-inflammatory cytokine IL-1β may downregulate DNMT3B in mouse and human chondrocytes via the NFκB (70). Long-term repetitive stimulation of primary generational chondrocytes with inflammatory cytokines induced demethylation of specific CpG sites in the proximal promoter of IL-1β, resulting in an increased and sustained IL-1β expression (71). The expression of inflammatory factors is upregulated, which in turn leads to an imbalance in joint cartilage metabolism through the NFκB pathway. The increase in cartilage degrading enzymes in advanced OA may be due to epigenetic changes in the methylation state of CpG sites in the promoter regions of these enzymes (72). In OA, numerous chondrocytes undergo phenotypic changes and acquire gene expression banks characterized by abnormal expression of numerous catabolic genes, including MMPs, aggregation enzymes (ADAMTS-4 and -5), iNOS, IL-1β and other cytokines (73,74). These pro-inflammatory cytokines work together with abnormally expressed cartilage catabolic genes to accelerate degradation of chondrocytes. Aging MSCs likewise produce numerous pro-inflammatory factors (SASPs) and may therefore affect OA by influencing DNA methylation (Fig. 2).
Histone modification
Histones are highly conserved proteins whose function is to stabilize, organize and concentrate DNA within a limited range of the nucleus. They consist of duplicate octamers containing dimers of each protein in four core histones (H2A, H2B, H3 and H4) and encapsulate the genomic DNA on its outer surface (66). Acetylation neutralizes the positive charge on the histone by oxidizing the amine residue into an amide and reduces the histone's ability to bind to DNA; this prevents chromatin shrinkage and allows the gene transcription machinery to enter the underlying DNA and transcribe. Deacetylation presents a positively charged histone tail, promoting high-affinity binding between the DNA backbone and histone, resulting in chromatin condensation, thereby blocking transcription (66).
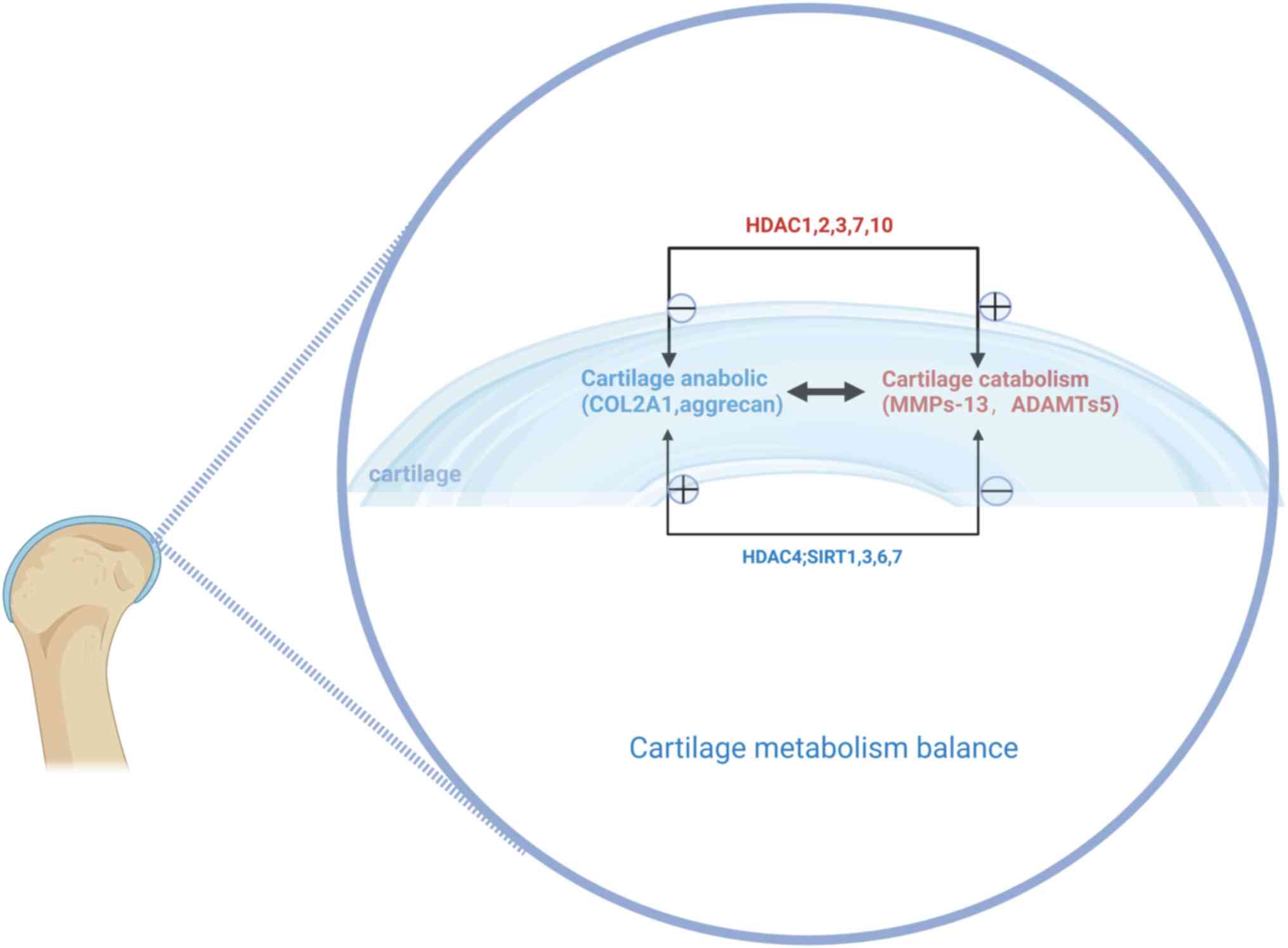
HDACs constitute a family of enzymes responsible for histone and non-histone deacetylation, whereas histone acetyltransferase catalyzes the reverse reaction, that is, acetylation. HDACs catalyze the removal of the acetyl functional groups from lysine residues of histones and non-histones. HDAC enzymes are divided into four classes: Class I Rpd3-like proteins (HDAC1, HDAC2, HDAC3 and HDAC8); class II Hda1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10); class III Sir2-like proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRRT7); and class IV proteins (HDAC11) (75). Studies have shown elevated levels of HDAC1 and HDAC2 in chondrocytes and synovial membranes of patients with OA than in the control group; the new carboxyl-terminal domain of HDAC1 and HDAC2 worked in tandem with the transcriptional inhibitor snail 1 to inhibit the expression of the collagen α1(II) gene (COL2A1) and aggrecan (76,77). Knockdown of human chondrocyte HDAC3 led to upregulated expression of cartilage oligomeric matrix protein (COMP), COL2A1, SOX9 and aggrecan, and downregulated expression of COL10 (78), whose overexpression may contribute to OA progression. miR-193b promoted cartilage differentiation of hMSCs by inhibiting HDAC3 expression, thereby maintaining cartilage-specific gene expression (78). Studies have shown that exogenous HDAC4 reduces the transcription of RUNX2, MMP1, MMP3, MMP-13, X-collagen, Indian hedgehog signal and ADAMTS-4 and -5, and increases the transcription of COL2. Furthermore, overexpression of HDAC4 not only reduced the expression of IL-1β, COX2 and iNOS and increased the expression of aggrecan, but also partially blocked the effect of IL-1β on the catabolic events in human OA chondrocytes (79); therefore, it may slow down the progress of OA. Studies have shown that high cartilage HDAC7 expression levels in human OA may lead to cartilage degradation by promoting MMP-13 gene expression (80). IL-1β upregulated IL-6 and IL-8 expression in synovial MSCs (SMSCs) through the NFκB pathway, and HDAC10 overexpression promoted the IL-6, IL-8 and IL-1β-mediated activation of the NFκB pathway (81). HDAC10 upregulation contributes to the activation of IL-1β-mediated SMSC inflammation (81), induces cartilage catabolic gene expression and causes cartilage metabolism imbalance. A decrease in the expression of HDACs can be observed in senescent MSCs (82). These studies suggested that aging MSCs may affect cartilage catabolism by influencing expression of HDAC1, 2, 3, 7 and 10, promoting OA progression, and HDAC4 helps mitigate this process (Fig. 3).
Sirtuins are a class of NAD-dependent HDACs associated with OA pathology
SIRT1-SIRT7 constitute NAD-dependent class III HDACs (83). Together with SOX9, SIRT1 has been shown to regulate the expression of the collagen gene COL2A1 in human chondrocytes (83,84). SIRT1 expression decreased with OA progression, and a decrease in SIRT1 in chondroblasts may lead to chondrocyte mast and cartilage matrix loss (85). Knocking down SIRT1 in young MSCs induces cellular senescence and inhibits cell proliferation, while upregulation of SIRT1 attenuates the biological senescence process, quenches age-related MSCs senescence (86). SIRT1 expression decreases in aging MSCs (86), which indirectly reflects that MSCs aging is associated with OA. Similarly, SIRT3 is an NAD+-dependent deacetylase, the expression of which is downregulated in senescent human MSCs (87). SIRT-3 mediated age-related changes in cartilage redox regulation and prevented early OA (88). Compared with healthy individuals, patients with OA have significantly lower levels of SIRT6 in articular chondrocytes (89). SIRT6 knockdown significantly inhibits the expression of genes in the extracellular chondrocytes (for example, COL2A1) and anabolic growth factors (for example, IGF-1) (90). In addition, SIRT6 knockdown makes human bone marrow MSCs aging (91). Overexpression of Sirt6 significantly inhibits NFκB-dependent transcriptional activity and prevents IL-1β-induced chondrocytes in OA (89). Sirt6 can also attenuate chondrocytes senescence by inhibiting IL-15/JAK3/STAT5 signaling (92). Thus, aging MSCs may affect cartilage metabolism by influencing SIRT6 expression. IL-1β in OA cartilage may downregulate levels of Sirt7 expression compared with healthy cartilage, and Sirt7 deficiency accelerates the catabolism of collagen II (93). Studies have shown that SIRT7 expression decreases during hematopoietic stem cell (HSCs) aging (94), and SIRT7 activation can improve the regenerative ability of senile HSCs (95). SIRT7 expression decreases during the aging of hMSCs, and SIRT7 deficiency may accelerate aging (96). Therefore, the expression of SIRT1, 3, 6 and 7 in aging MSCs is reduced, affecting cartilage breakdown anabolism, which in turn promotes the development of OA.
Histone methylation and acetylation are also important epigenetic regulators during cartilage differentiation (65,97). Similar to DNA, histones can be methylated by histone methyltransferase and demethylated by histone demethylase, both of which can alter gene transcription (66). Decreased expression of HDACs is observed in senescent MSCs (82). Decreased H3K9 methylation increased the transcription of MMP-1 and MMP-13 in chondrocytes. Transcription of the anabolic factors SOX9 and COL2A1, which are involved in cartilage differentiation, were also reduced by treatment with histone methyltransferase inhibitor chaetocin (98). Therefore, histone methylation contributes to cartilage differentiation. Lysine demethylase 6B (Kdm6b), also known as Jmjd3, has been identified as an H3K27 demethylase that catalyzes H3K27me2/3 demethylation; knockdown of Kdm6b in chondrocytes led to abnormal cartilage development and accelerated OA progression by inhibiting the anabolic metabolism of chondrocytes (65). Kdm6b promoted chondroblast proliferation and hypertrophy during intrachondral osteogenesis in Kdm6b−/− mice (99). Depletion of Kdm6A inhibited the expression of SOX9, COL2A1 and ACAN, resulting in an increase in H3K27me3 and a decrease in H3K4me3 levels (100). Similarly, KDM4B is a histone demethylase that mediates transforming growth factor (TGF)-induced SOX9 activation by removing H3K9me3 from the SOX9 promoter (101). Both KDM4B and KDM6B promoted osteogenesis differentiation of human MSCs (102). The pro-inflammatory cytokine TGF-β1 promotes upregulation of KDM4B and KDM6B (103). Thus, senescent MSCs can affect articular cartilage metabolism and OA by releasing SASPs, affecting histone methylation.
Other non-coding RNAs
The main types of non-coding RNAs include microRNAs (miRNAs), lncRNAs, and circular RNAs (circRNAs) (104). Non-coding RNA is divided into small RNA (<200 nucleotides) and long RNA (>200 nucleotides).
miRNA
The most well-known subset of small RNA is miRNAs, which are 18-25 nucleotides (105). miRNA expression is influenced by pro-inflammatory cytokines. Multiple studies have shown that certain non-coding RNAs, especially miR-140, are involved in the development of OA by modulating key catabolic factors. miR-140 is a conserved miRNA that is important for both chondrogenesis and osteogenesis and is usually highly expressed in normal cartilage. miR-140 contributes to joint cartilage development and homeostasis as well as normal intrachondral bone development (106,107). Previous studies showed reduced miR-140 expression in human OA cartilage (108,109). Treatment of chondrocytes with IL-1β in vitro inhibited miR-140 expression. By contrast, miR-140-transfected chondrocytes downregulated IL-1β-induced ADAMTS5 (polymerase degrading enzyme 5) expression (107,109). Thus, miR-140 can affect OA by affecting articular cartilage anabolic catabolism. Similar to miR-140, miR-30a directly targeted ADAMTS5, whereas IL-1β inhibited miR-30a expression by activating activator protein 1 (c-jun/c-fos), thereby upregulating ADAMTS5 expression; thus, the downregulation of miR-30a in OA is negatively correlated with ADAMTS5 expression (110). miR-92a-3p, which is involved in advanced chondrogenesis of hMSCs, was downregulated in human OA cartilage and responded to IL-1β in vitro (111). Mao et al (111,112) studied the effects of human MSC (hMSC)-derived exosomes overexpressing miR-92a-3p on cartilage formation and cartilage degeneration and found that the exosomes increased the transcription and translation of aggrecan, COL2A1 and SOX9, and reduced the expression levels of COL10A1, RUNX2 and MMP-13. IL-1β mediated miR-92a-3p downregulation in human OA cartilage, which may promote aggrecan degradation in OA (112); this exacerbated cartilage destruction. Expression of miR-193b-3p in degenerative cartilage was significantly reduced; IL-1β-induced downregulation of miR-193b-3p led to a significant increase in HDAC3, COL10A1 and MMP-13 with corresponding decreases in SOX9, COL2A1, COMP and aggrecan expression (78). miR-105 downregulation, through recruitment of promoter NFκB/p65 complexes, led to an upregulation of ADAMTS by targeting RUNX2 (58). IL-1β-induced downregulation of miR-27b led to increased MMP-13 expression (113). These studies demonstrated that IL-1β stimulation downregulated these miRNAs, resulting in corresponding cartilage-specific gene downregulation and cartilage catabolic gene upregulation, thereby influencing OA progression (Fig. 4).
Aging MSCs cause the downregulation of the corresponding miRNA by producing SASP, thus affecting cartilage metabolism. Other miRNAs also affect OA through various mechanisms, such as miR-145, which inhibited cartilage differentiation of TGF-β3-induced MSCs by directly targeting SOX9 at the post-transcriptional level, resulting in its downregulation (114). miR-146a antagonized the expression of IL-1-induced cartilage degrading enzymes MMP-13 and ADAMTS5 in articular cartilage (115). Overexpression of miR-148a increased COL2A1 expression and decreased COL10A1, MMP-13 and ADAMTS5 gene expression (116). Inhibition of miRNA-495 relieved IL-1β-induced chondroblastic inflammatory response by salvaging SOX9 expression (117). miR-129-5p decreased in patients with OA and IL-1β-induced chondrocytes; miR-129-5p in exosomes from human SMSC (hSMSC-Exo) inhibited IL-1β-mediated OA by inhibiting the release of high mobility group box protein 1 (HMGB1) (118). Overexpression of miRNA-615-3p led to increased expression levels of inflammatory cytokines such as IL-1, IL-6 and IL-α (119). Therefore, miRNA-615-3p overexpression may also affect articular cartilage metabolism through the NF-kB pathway. Human bone marrow MSCs (hBMSCs) exhibited typical MSC differentiation potential. During cartilage differentiation, the expression of collagen 2 and 10 (COL2 and COL10), SOX9, and RUNX2 are upregulated. Expression levels of miR-140, miR-143 and miR-181a increased over time, whereas those of miR-27b, miR-221 and miR-615-3p decreased (120) during cartilage differentiation in BMSC. miR-26b expression was significantly downregulated during the in vitro cartilage differentiation of rat MSCs, and it played an inhibitory role in the process by inhibiting Wnt expression (121). miR-485-5p levels were inversely correlated with the degree of differentiation of BMSCs; miR-485-5p reduced SOX9 levels, promoted the synthesis of cartilage surface inflammatory factors, and blocked mouse BMSCs from differentiating into chondrocytes (122). miR-539-3p is gradually downregulated during cartilage differentiation of human adipose-derived MSCs (hADMSCs); its overexpression inhibited cartilage differentiation of TGF-β1-induced hADMSCs by reducing gene and protein expression of cartilage differentiation markers COL2A1 and ACAN, with SOX9 being a direct target gene for miR-539-3p. During cartilage differentiation of hADMSCs, the expression of SOX9 is gradually upregulated over time (123). miR-210-3p promotes cartilage differentiation of rat BMSCs and promotes mRNA and protein levels of cartilage expression genes COL2 and SOX9 (124). miR-130b inhibitors induce chondrocyte differentiation and chondrocyte growth of BMSCs by targeting SOX9 (125). Long intergenic non-protein coding RNA, a regulator of reprogramming (Linc-ROR), is downregulated in the articular cartilage tissue of patients with OA; it promotes cartilage differentiation and cartilage formation by BMSCs by acting as competitive endogenous RNAs for miR-138 and miR-145 and activating SOX9 expression (126). A previous study showed overexpression of miR-122-5p by indolol 2,3 dioxygenase 1 in synovial fluid in patients with OA, activating the Wnt1/β-catenin pathway to impair cartilage differentiation and cartilage regeneration of MSCs (127). ThesemiRNA changes induce mesenchymal stem cell cartilage differentiation through various pathways (Fig. 4). Senescent MSCs release pro-inflammatory cytokines that mediate cartilage damage and apoptosis and can partially alleviate cartilage damage by modulating miRNAs expression.
LncRNA
LncRNAs can be divided into small (200-950 nt), medium (950-4,800 nt), and large lncRNAs (~4,800 nt) (128), and are rich in MSC-derived exosomes. Numerous lncRNAs regulate the chondrogenesis of MSCs. For example, lncRNA ZBED3-AS1 induced chondrogenesis of MSC of human synovial fluid origin (SFMSCs) (129). LncRNA ROCR contributed to SOX9 expression and cartilage differentiation of hMSC (47). LncRNA GRASLND (originally named RNF144A-AS1) acted as a regulator of MSC chondrogenesis (130). LncRNA DANCR was involved in SFMSC proliferation and chondrogenesis. Mechanically, DANCR acted as a sponge RNA of miR-1275, regulating the expression of the target gene MMP-13 (131). The expression of lnc-RNA BLACAT1 in inflammatory BMSCs increased, and the knockdown of BLACAT1 promoted proliferation and osteogenesis differentiation of BMSCs targeting miR-142-5p (132). LncRNA XIST silencing promoted cartilage differentiation of SMSCs. In addition, XIST regulated the expression of ADAMTS-5 by directly binding to miR-27b-3p (133). TIMP-3 (a natural inhibitor of MMPs) expression may be downregulated by recruiting DNMT1, DNMT3A and DNMT3B, thereby increasing the methylation ratio of CpG islands in the promoter region of TIMP-3, thereby increasing collagen degradation in OA chondrocytes (134). MSC-derived exosomes (MSC-exo)-mediated lncRNA KLF3-AS1 inhibited autophagy and apoptosis of IL-1β-treated chondrocytes via the PI1K/Akt/mTOR signaling pathway (135). Exosome lncRNA-KLF3-AS1 from hMSCs is significantly enriched in MSC-exo, which improves IL-1β-induced cartilage damage; exosome lnc-KLF3-AS1 inhibited IL-1β-induced chondrocyte apoptosis (136). Overexpression of lncRNA CTBP1-AS2 downregulated miR-130a by increasing methylation levels of the miR-130a gene, ultimately leading to a decrease in the rate of chondrocyte proliferation in patients with OA (137). These studies have shown that lncRNAs can regulate chondrogenesis of MSCs, and some lncRNAs can regulate the expression of cartilage-specific genes and catabolic genes through various mechanisms. SASP produced by aging MSCs induces cartilage damage and apoptosis, aggravating OA. Numerous lncRNAs can delay OA progression by regulating cartilage formation of MSCs.
circRNA
Cyclic RNAs (circRNAs) are a new class of discovered non-coding RNAs with structural stability. For example, Circ_ATRNL1 is significantly higher in cartilage tissues of patients with OA. Circ_ATRNL1 overexpression enhanced proliferation and differentiation of hADMSCs into chondrogenesis, promoted the expression of COL2, ACAN and SOX9, and inhibited the adipose differentiation of hADMSC and the expression of adipose-related genes. miR-145-5p is a target miRNA for circ_ATRNL1 and SOX9 (138). A miR-145-5p mimic inhibited the differentiation of cartilage by hADMSC and the expression of cartilage-related factors. The miR-145-5p mimic effectively reversed the regulatory effect of circ_ATRNL1 on hADMSC. Circ_ATRNL1 upregulated SOX9 expression to promote cartilage differentiation of hADMSCs mediated by miR-145-5p (138). MSCs-circHIPK3-EV [extracellular vesicles (EV) from MSCs overexpressing circHIPK3] significantly improved IL-1β-induced chondrocyte damage (139). Similar to lncRNA and miRNAs, circRNAs can also alleviate OA by modulating the expression of cartilage-specific genes and catabolic genes. Therefore, by regulating the expression of non-coding RNA, it affects the expression of cartilage-specific genes (COL2A1 and aggrecan) and cartilage catabolic genes (MMP-13 and ADAMTS5) in OA.
In summary, epigenetic changes such as decreased DNA methylation, histone acetylation and methylation, and regulation of non-coding RNA greatly affect the articular cartilage metabolism in OA. Epigenetic changes that occur in senescent cells can also affect joint cartilage metabolism. In the future, the development of therapies targeting cellular senescence is expected to become a specific treatment for OA.
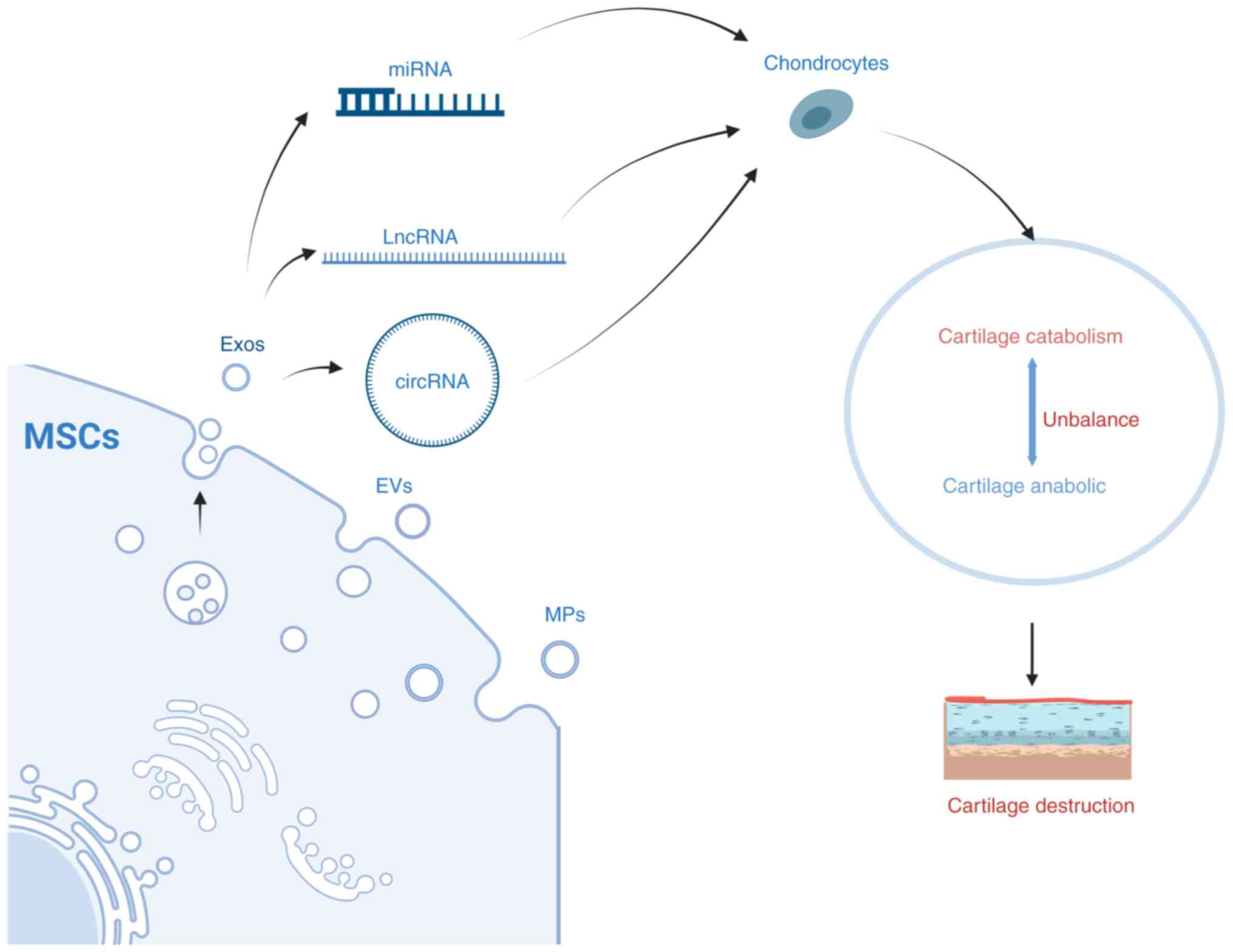
6. Exosomes from MSCs are modified into various miRNAs that influence OA progression
Exosomes are tiny membrane-bound vesicles released from cells. Exosomes from different types of MSCs, including BMSCs, SMSCs, ADMSCs and embryonic MSCs (EMSCs) regulated cartilage regeneration and slowed OA progression, whereas exosomes isolated from the synovial fluid stimulated the release of inflammatory cytokines and MMPs (140).
MSC-EV promoted the expression of Col2A1, sox9 and Acan in mouse chondrocytes in OA models, while negatively regulating the expression of Mmp-13 and Runx2. MSC-circHIPK3-EV significantly improved IL-1β-induced chondrocyte damage (139), and MSC-KLF3-AS1-exo (exosomes from KLF3-AS1-overexpressed-MSCs) improved IL-1β-induced chondrocyte damage by participating in MSC-mediated chondrocyte proliferation induction and chondrocyte apoptosis inhibition via the miR-206/G protein-coupled receptor kinase interaction protein-1 axis (141). Expression of exosome miR-92a-3p in MSC cartilage exosomes is elevated, whereas that in exosomes secreted by OA chondrocytes is significantly reduced. MSC-miR-92a-3p-exo promoted cartilage proliferation and matrix gene expression in MSCs and OA primary human chondrocytes (PHCs), respectively. Conversely, treatment with MSC-anti-miR-92a-3p-exo inhibited cartilage differentiation and reduced cartilage stromal synthesis by enhancing the expression of WNT5A (142).
Exosomes from hBMSCs overexpressed miR-92a-3p and directly targeted the WNT5A gene to increase the expression of chondrocyte markers (for example, COL2 and SOX9) and reduce catabolic markers (for example, MMP-13 and RUNX2), thereby increasing the proliferation of chondrocytes and protecting chondrocytes from apoptosis (142). hBMSC-derived exosome-metastatic miR-361-5p mitigated chondrocyte damage and inhibited the NFκB signaling pathway by targeting DDX20. Inhibition of NFκB signaling reversed the effect of overexpressed DDX20 on IL-1β-induced chondrocyte damage. In addition, the exosome miR-361-5p reduced OA damage in vivo (143). Similarly, BMSC-derived exosomes and microvesicles/microparticles re-induced the expression of chondrocyte markers (COL2 and ACAN) while inhibiting catabolism (MMP-13 and ADAMTS5) and inflammatory (iNOS) markers (144). Thus, inhibiting IL-1β-induced chondrocytes destruction. BMSC-derived exosomes are rich in miR-125a-5p, which is endowed with features that accelerate chondrocyte migration; this is accompanied by a higher expression of COL2, ACAN and SOX9 and lower expression of MMP-13 in vitro, and also relieve ECM degradation of chondrocytes (145). In clinical samples of traumatic OA cartilage tissue, ELF3 expression increased and miR-136-5p expression decreased. BM-MSC-exo showed high levels of miR-136-5p, which was internalized by chondrocytes. miR-136-5p promoted the migration of chondrocytes, wherein the expression of COL2, ACAN and SOX9 increased and that of MMP-13 decreased, verifying that miR-136-5p targeted ELF3 and downregulated its expression (146). Human ADMSC-EV not only promoted the proliferation and migration of human OA chondrocytes, but also maintained the chondrocyte matrix in the presence of IL-1β by increasing COL2 synthesis and reducing the expression of MMP-1, MMP-3, MMP-13 and ADAMTS-5 (147). Exosomes derived from SMSCs overexpressing miR-155-5p promoted the proliferation and migration of OA chondrocytes, inhibited apoptosis and enhanced ECM secretion, thus effectively blocking OA in mouse models (148). In addition, the SMSC-EV carried miR-26a-5p into chondrocytes, upregulated miR-26a-5p, and inhibited phosphatase and tension homologs, thereby inhibiting apoptosis and inflammation and improving cartilage damage in OA (149). SMSC-EV mitigated chondrocyte damage during OA by miR-130b-3p-mediated inhibition of the LRP12/AKT/β-catenin axes (150). miR-129-5p in hSMSC-Exo alleviated IL-1β-mediated OA by inhibiting the release of HMGB1, while downregulating the expression of iNOS, COX2, MMP-13, and NFκB (118). The SMSC-derived exosome miR-320c enhanced chondrogenesis by targeting ADAM19, highlighting the potential new mechanism of SMSC in the treatment of OA (151). Co-culture of ADMSCs and SMSCs with chondrocytes reduced MMP-13 expression, while increasing the expression of COL2A1, ACAN and SOX9, and thus, reversed the IL-1β effect on promoting reactive oxygen species content and inflammatory factor levels (152). EMSC-exo also had a beneficial therapeutic effect on OA by increasing the expression of COL2 in the cartilage matrix and reducing the expression of ADAMTS5 (153). Exosome overexpression of miR-140-5p from human urine-derived stem cells enhanced cartilage regeneration and subchondral bone remodeling (154). Exosomes from miR-95-5p-overexpressing primary chondrocytes (AC-miR-95-5p) enhanced chondrogenicity and prevented the development of OA by directly targeting HDAC2/8 (155). The role of exosomes in OA discussed in this review is shown in (Table I).
The aforementioned studies have shown that MSCs-derived exosomes improve OA by regulating cartilage development and cartilage injury repair through mechanisms such as altering miRNA, lncRNA and circRNA expression (Fig. 5). This has become a research hotspot in recent years. There is growing evidence that exosomes can effectively reduce inflammation, promote cartilage regeneration and reduce pain in OA patients. In the future, the potential of exosomes in the treatment of OA should be used to a full extent towards the development of novel specific exosome therapies.
7. Single-cell sequencing reveals the mechanism by which the aging MSCs affect OA
Single-cell RNA sequencing (scRNA seq) assesses individual cells, identifies new cell populations, reveals regulatory relationships between genes and tracks the trajectory of different cell lineages in development and even disease (156). This contributes to a comprehensive understanding of the pathophysiology of the disease and facilitates the development of effective treatment options. In terms of cartilage development, scRNA seq helps identify self-renewing and pluripotent human bone stem cells that produce bone, cartilage and stromal progenitor cells (157). Previous studies identified four chondrocytes subclusters including proliferative chondrocytes (ProCs), pre-hypertrophic chondrocytes (preHTCs), HTCs and fibrochondrocytes (FCs). HDAC4 expressed in preHTCs regulates chondrocytes hypertrophy and endochondral bone formation by interacting with the transcription factor Runx2, which is necessary for chondrocytes hypertrophy, and inhibiting its activity (158). Overexpression of HDAC4 in ProCs in vivo inhibits chondrocytes hypertrophy and differentiation (158). HDACs expression decreased in aging MSCs (82). Therefore, aging MSCs may promote chondrocytes to hypertrophy and aggravate OA. HTCs attract blood vessels and osteocytes to invade and then undergo apoptosis and calcium deposition, ultimately triggering the progression of OA in humans (159). FCs are mainly present in the late stages of OA and have a high proportion of genes and angiogenesis ability associated with adverse OA outcomes, indicating that FCs promote OA progression (159). Previously, a study using scRNA seq on OA cartilage identified three new subsets of cartilage in cartilage, effector chondrocytes (ECs), regulatory chondrocytes (RegCs) and homeostatic chondrocytes (HomCs), and highlighted some changes associated with OA progression (159). ECs are associated with the tricarboxylic acid cycle and amino acid metabolism, suggesting that they provide cellular energy (159). Another study showed that ECs mainly exert immune function, causing tissue inflammation, and possibly promoting the development of OA (160). HomCs exhibit high expression of genes related to cell cycle regulation, metabolic processes and development in response to external stimuli (159,161). RegCs are involved in numerous signaling pathways and may play a key role in regulating OA progression (159). In addition, Ji et al (159) identified two hypertrophic chondrocytes subclusters: HTC-A is highly expressed in genes associated with cartilage development and connective tissue development; HTC-B is highly expressed in genes associated with ECM tissue, ossification and mineralization. In 2021, Sebastian et al (162) identified nine chondroblast subtypes (Ucmahigh, Cytl1high, Chil1high, Mef2chigh, Krt16high, Tnfaip6high, S100a4high, Neat1high and divC chondrocyte clusters) in the knee cartilage of healthy mice by analyzing cells from each cluster separately. MMP-3, Inhba, Sfn, Il11, Ptgs2, Dusp2 and MMP-13 are upregulated among various chondrocytes subtypes, while Cytl1, Il17b, Fgfr2, Ptch1, Dbp and Rad are downregulated. The Ucmahigh and Krt16high clusters have unique ECM signatures with increased expression of several key ECM proteins, including COL2A1, COL9A1-a3 and ACAN (162). Previous studies have found elevated expression of Mmp3, Mmp13, Ptgs2, Il11 and Inhba in OA (163,164). The results of Sebastian et al (162) confirmed that joint chondrocytes are the main source of this elevated expression in injured joints. SPP1 was found to be highly expressed in OA cartilage, and Qu et al (165) identified unique chondrocytes characterized by high SPP1 expression. Gao et al (166) performed scRNA seq on human articular chondrocytes treated with IL-1β and it found that one of the initial cell clusters can be transformed into a pro-inflammatory subpopulation through a pathway called an inflammatory response, which may be a central target for mitigating OA progression. Yoshimoto et al (167) demonstrated by analyzing scRNA seq datasets that cellular senescence signals in chondrocytes are associated with OA pathogenesis. The emergence of scRNA seq has broadened our understanding of chondrocytes subsets. scRNA seq can also play an important role in exploring the association between MSC aging and OA pathogenesis. In the future, improved identification of the functional roles of subpopulations and the description of relevant molecular mechanisms are needed in order to improve understanding of the pathogenesis of OA, identify new therapeutic targets and develop specific therapies.
8. Highlights
The present review mainly explored the relationship between mesenchymal stem cell senescence and genetic and epigenetic changes in the progression of OA from the perspective of pro-inflammatory cytokines. OA progression is associated with genetic and epigenetic changes that occur during mesenchymal stem cell aging.
Non-coding RNA influences OA progression by modulating cartilage synthesis/catabolism.
The exosomes secreted by various mesenchymal stem cell sources regulate miRNA expression to affect metabolism in joint cartilage. Exosome treatment for OA is a promising future direction.
9. Summary
OA is a disease associated with age and cartilage destruction. Pro-inflammatory cytokines play a pivotal role in the development of OA. Cytokines activate white blood cells, which produce more cytokines. Therefore, even a small pro-inflammatory stimulus may produce a more systemic chronic inflammatory response. Spontaneous differentiation and aging of MSCs occur during expansion, and little is currently known about the molecular mechanisms involved. Aging MSCs release pro-inflammatory cytokines that cause an inflammatory storm and aggravate the progression of OA. More studies are needed to determine the molecular mechanisms of MSCs aging to identify new specific targets and promote development of specific OA therapies. With the application of large-scale GWAS, the role of OA genetics in OA is gradually emerging. The proportion of known OA risk loci in heritability is just >20%, and there are still a large number of loci to be discovered. Interpreting the results of GWAS is biologically challenging, therefore translating genetic discoveries into effective treatments remains elusive for now. In the future, as more GWAS sites are reported, the gap between discovery and utility will narrow. Epigenetics is a key player in the formation and maintenance of joints. OA is associated with alterations in numerous epigenetic markers in the affected tissues, such as DNA methylation, histones and non-coding RNA. Exosome therapy for OA has shown exciting promise over the past few years, with MSC-derived exosomes becoming potential cell-free therapies for OA because of their anti-inflammatory properties as well as unique advantages and characteristics. The therapeutic effect of MSC-derived exosomes is similar to that of MSCs. Numerous studies have demonstrated that MSC-derived exosomes mediate cartilage tissue repair, promote cartilage differentiation of MSCs, and attenuate the development of OA; this suggests that MSC-derived exosomes have exciting therapeutic potential for OA. However, the specific signaling molecules that regulate MSC-derived exosomes in OA are still not well understood. Clarifying the underlying molecular mechanisms of MSC-derived exosomes in OA pathogenesis will help identify potential therapeutic targets for OA. Despite the great potential of exosomes in the treatment of OA, obtaining consistent and abundant high-quality exosomes remains a technical challenge. Obtaining high-quality exosomes maximizes their effectiveness and ensures their safe application. Single-cell sequencing has shone in recent years in identifying chondrocytes subsets; however, the understanding of the molecular mechanisms of orthopedic disease development and homeostasis by single-cell sequencing is still in infancy. In the future, attention should be paid to identify subsets or disease-specific subsets, key signaling pathways and conduct in-depth research on the functions of subpopulations and their interactions to improve understanding of OA. Meanwhile, scRNA-seq could be combined with other traditional and emerging technologies to complement each other to achieve more detailed and accurate cell classification and expand our understanding of orthopedic diseases.
Availability of data and materials
Not applicable.
Authors' contributions
DT and ZH wrote the manuscript. ZZ, ZD and WL designed and corrected the whole manuscript. DW, XC and JL proposed the information. DW supervised the process. ZD and WL funded the project. All authors read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81800785, 81972085 and 82172465), the China University Industry-University-Research Innovation Fund (grant no. 2021JH037), the Natural Science Foundation of Guangdong (grant nos. 2018A0303100027 and 2021A1515010706), the Guangdong Provincial Key Clinical Discipline-Orthopedics (grant no. 2000005), the Sanming Project of Shenzhen Health and Family Planning Commission (grant no. SZSM201612086), the Shenzhen Science and Technology Planning (grant nos. JCYJ20180228163401333 and JCYJ20190806170612680), the Shenzhen Key Medical Discipline Construction Fund (grant no. SZXK025), the discipline construction Capacity Improvement project of Shenzhen Municipal Health Commission (grant no. SZXJ2018065) and the Natural Science Foundation of Guangdong (grant no. 2023A1515010102).
References
|
Loeser RF, Goldring SR, Scanzello CR and Goldring MB: Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 64:1697–1707. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Abramoff B and Caldera FE: Osteoarthritis: Pathology, diagnosis, and treatment options. Med Clin North Am. 104:293–311. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Lan T, Luo M and Wei X: Mesenchymal stem/stromal cells in cancer therapy. J Hematol Oncol. 14:1952021. View Article : Google Scholar : PubMed/NCBI | |
|
Hernandez-Segura A, Nehme J and Demaria M: Hallmarks of cellular senescence. Trends Cell Biol. 28:436–453. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
López-Otín C, Blasco MA, Partridge L, Serrano M and Kroemer G: The hallmarks of aging. Cell. 153:1194–1217. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Xu M, Bradley EW, Weivoda MM, Hwang SM, Pirtskhalava T, Decklever T, Curran GL, Ogrodnik M, Jurk D, Johnson KO, et al: Transplanted senescent cells induce an osteoarthritis-like condition in mice. J Gerontol A Biol Sci Med Sci. 72:780–785. 2017. | |
|
Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY and Campisi J: Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6:2853–2868. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Xu M, Tchkonia T, Ding H, Ogrodnik M, Lubbers ER, Pirtskhalava T, White TA, Johnson KO, Stout MB, Mezera V, et al: JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc Natl Acad Sci USA. 112:E6301–E6310. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Greene MA and Loeser RF: Aging-related inflammation in osteoarthritis. Osteoarthritis Cartilage. 23:1966–1971. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Lotz M and Loeser RF: Effects of aging on articular cartilage homeostasis. Bone. 51:241–248. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, Chung JW, Kim DH, Poon Y, David N, et al: Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 23:775–781. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ding DC, Shyu WC and Lin SZ: Mesenchymal stem cells. Cell Transplant. 20:5–14. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Wu Q, Wang Y, Li L, Bu H and Bao J: Senescence of mesenchymal stem cells (Review). Int J Mol Med. 39:775–782. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Alt EU, Senst C, Murthy SN, Slakey DP, Dupin CL, Chaffin AE, Kadowitz PJ and Izadpanah R: Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. 8:215–225. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Chew JRJ, Chuah SJ, Teo KYW, Zhang S, Lai RC, Fu JH, Lim LP, Lim SK and Toh WS: Mesenchymal stem cell exosomes enhance periodontal ligament cell functions and promote periodontal regeneration. Acta Biomater. 89:252–264. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Loeser RF, Collins JA and Diekman BO: Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 12:412–420. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Lin Z, Deng Z, Liu J, Lin Z, Chen S, Deng Z and Li W: Chloride channel and inflammation-mediated pathogenesis of osteoarthritis. J Inflamm Res. 15:953–964. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Xu X, Liang Y, Li X, Ouyang K, Wang M, Cao T, Li W, Liu J, Xiong J, Li B, et al: Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid-derived mesenchymal stem cells and cartilage regeneration. Biomaterials. 269:1205392021. View Article : Google Scholar | |
|
McCulloch K, Litherland GJ and Rai TS: Cellular senescence in osteoarthritis pathology. Aging Cell. 16:210–218. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Duan L, Liang Y, Xu X, Xiao Y and Wang D: Recent progress on the role of miR-140 in cartilage matrix remodelling and its implications for osteoarthritis treatment. Arthritis Res Ther. 22:1942020. View Article : Google Scholar : PubMed/NCBI | |
|
Fujii Y, Liu L, Yagasaki L, Inotsume M, Chiba T and Asahara H: Cartilage homeostasis and osteoarthritis. Int J Mol Sci. 23:63162022. View Article : Google Scholar : PubMed/NCBI | |
|
Liu CF and Lefebvre V: The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 43:8183–8203. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al: Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 434:644–648. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im HJ and Chen D: MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res Ther. 15:R52013. View Article : Google Scholar : PubMed/NCBI | |
|
Philipot D, Guérit D, Platano D, Chuchana P, Olivotto E, Espinoza F, Dorandeu A, Pers YM, Piette J, Borzi RM, et al: p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res Ther. 16:R582014. View Article : Google Scholar : PubMed/NCBI | |
|
Loeser RF: Aging and osteoarthritis: The role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis Cartilage. 17:971–979. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP and Fahmi H: Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 7:33–42. 2011. View Article : Google Scholar | |
|
Fang H, Deng Z, Liu J, Chen S, Deng Z and Li W: The mechanism of bone remodeling after bone aging. Clin Interv Aging. 17:405–415. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Bian Q, Wang YJ, Liu SF and Li YP: Osteoarthritis: Genetic factors, animal models, mechanisms, and therapies. Front Biosci (Elite Ed). 4:74–100. 2012. View Article : Google Scholar | |
|
Molnar V, Matišić V, Kodvanj I, Bjelica R, Jeleč Ž, Hudetz D, Rod E, Čukelj F, Vrdoljak T, Vidović D, et al: Cytokines and chemokines involved in osteoarthritis pathogenesis. Int J Mol Sci. 22:92082021. View Article : Google Scholar : PubMed/NCBI | |
|
Hwang HS and Kim HA: Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 16:26035–2604. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Deng Z, Chen X, Lin Z, Alahdal M, Wang D, Liu J and Li W: The homeostasis of cartilage matrix remodeling and the regulation of volume-sensitive ion channel. Aging Dis. 13:787–800. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Chen LX, Lin L, Wang HJ, Wei XL, Fu X, Zhang JY and Yu CL: Suppression of early experimental osteoarthritis by in vivo delivery of the adenoviral vector-mediated NF-kappaBp65-specific siRNA. Osteoarthritis Cartilage. 16:174–184. 2008. View Article : Google Scholar | |
|
Fei J, Liang B, Jiang C, Ni H and Wang L: Luteolin inhibits IL-1β-induced inflammation in rat chondrocytes and attenuates osteoarthritis progression in a rat model. Biomed Pharmacother. 109:1586–1592. 2019. View Article : Google Scholar | |
|
Huang X, Xi Y, Pan Q, Mao Z, Zhang R, Ma X and You H: Caffeic acid protects against IL-1β-induced inflammatory responses and cartilage degradation in articular chondrocytes. Biomed Pharmacother. 107:433–439. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Meszaros E and Malemud CJ: Prospects for treating osteoarthritis: Enzyme-protein interactions regulating matrix metalloproteinase activity. Ther Adv Chronic Dis. 3:219–229. 2012. View Article : Google Scholar | |
|
Verma P and Dalal K: ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. J Cell Biochem. 112:3507–3514. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Deng Z, Lin Z, Zhong Q, Lu M, Fang H, Liu J, Duan L, Chen L, Wang L, Wang D and Li W: Interleukin 1 beta-induced chloride currents are important in osteoarthritis onset: An in vitro study. Acta Biochim Biophys Sin (Shanghai). 53:400–409. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
oman-Blas JA and Jimenez SA: NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthritis Cartilage. 14:839–848. 2006. View Article : Google Scholar | |
|
Lepetsos P, Papavassiliou KA and Papavassiliou AG: Redox and NF-κB signaling in osteoarthritis. Free Radic Biol Med. 132:90–100. 2019. View Article : Google Scholar | |
|
Tang J, Cui W, Song F, Zhai C, Hu H, Zuo Q and Fan W: Effects of mesenchymal stem cells on interleukin-1β-treated chondrocytes and cartilage in a rat osteoarthritic model. Mol Med Rep. 12:1753–1760. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Wang T and He C: Pro-inflammatory cytokines: The link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 44:38–50. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Song H and Park KH: Regulation and function of SOX9 during cartilage development and regeneration. Semin Cancer Biol. 67:12–23. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Lefebvre V and Dvir-Ginzberg M: SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res. 58:2–14. 2017. View Article : Google Scholar : | |
|
Kawakami Y, Tsuda M, Takahashi S, Taniguchi N, Esteban CR, Zemmyo M, Furumatsu T, Lotz M, Izpisúa Belmonte JC and Asahara H: Transcriptional coactivator PGC-1alpha regulates chondrogenesis via association with Sox9. Proc Natl Acad Sci USA. 102:2414–2419. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Akiyama H, Stadler HS, Martin JF, Ishii TM, Beachy PA, Nakamura T and de Crombrugghe B: Misexpression of Sox9 in mouse limb bud mesenchyme induces polydactyly and rescues hypodactyly mice. Matrix Biol. 26:224–233. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Barter MJ, Gomez R, Hyatt S, Cheung K, Skelton AJ, Xu Y, Clark IM and Young DA: The long non-coding RNA ROCR contributes to SOX9 expression and chondrogenic differentiation of human mesenchymal stem cells. Development. 144:4510–4521. 2017.PubMed/NCBI | |
|
Nakamura Y, He X, Kato H, Wakitani S, Kobayashi T, Watanabe S, Iida A, Tahara H, Warman ML, Watanapokasin R and Postlethwait JH: Sox9 is upstream of microRNA-140 in cartilage. Appl Biochem Biotechnol. 166:64–71. 2012. View Article : Google Scholar | |
|
Yang J, Qin S, Yi C, Ma G, Zhu H, Zhou W, Xiong Y, Zhu X, Wang Y, He L and Guo X: MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 585:2992–2997. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Si HB, Zeng Y, Liu SY, Zhou ZK, Chen YN, Cheng JQ, Lu YR and Shen B: Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthritis Cartilage. 25:1698–1707. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang Y, Mishima H, Sakai S, Liu YK, Ohyabu Y and Uemura T: Gene expression analysis of major lineage-defining factors in human bone marrow cells: Effect of aging, gender, and age-related disorders. J Orthop Res. 26:910–917. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Kearns AE, Khosla S and Kostenuik PJ: Receptor activator of nuclear factor kappaB ligand and osteoprotegerin regulation of bone remodeling in health and disease. Endocr Rev. 29:155–192. 2008. View Article : Google Scholar | |
|
Li F and Li X, Liu G, Gao C and Li X: Bone marrow mesenchymal stem cells decrease the expression of RANKL in collagen-induced arthritis rats via reducing the levels of IL-22. J Immunol Res. 2019:84592812019. View Article : Google Scholar : PubMed/NCBI | |
|
Lin TH, Gibon E, Loi F, Pajarinen J, Córdova LA, Nabeshima A, Lu L, Yao Z and Goodman SB: Decreased osteogenesis in mesenchymal stem cells derived from the aged mouse is associated with enhanced NF-κB activity. J Orthop Res. 35:281–288. 217 | |
|
Wang X, Manner PA, Horner A, Shum L, Tuan RS and Nuckolls GH: Regulation of MMP-13 expression by RUNX2 and FGF2 in osteoarthritic cartilage. Osteoarthritis Cartilage. 12:963–973. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Chen D, Kim DJ, Shen J, Zou Z and O'Keefe RJ: Runx2 plays a central role in osteoarthritis development. J Orthop Translat. 23:132–139. 2019. View Article : Google Scholar | |
|
Li Z, Liu C, Xie Z, Song P, Zhao RC, Guo L, Liu Z and Wu Y: Epigenetic dysregulation in mesenchymal stem cell aging and spontaneous differentiation. PLoS One. 6:e205262011. View Article : Google Scholar : PubMed/NCBI | |
|
Ji Q, Xu X, Xu Y, Fan Z, Kang L, Li L, Liang Y, Guo J, Hong T, Li Z, et al: miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J Mol Med (Berl). 94:681–694. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Aubourg G, Rice SJ, Bruce-Wootton P and Loughlin J: Genetics of osteoarthritis. Osteoarthritis Cartilage. 30:636–649. 2022. View Article : Google Scholar : | |
|
Tachmazidou I, Hatzikotoulas K, Southam L, Esparza-Gordillo J, Haberland V, Zheng J, Johnson T, Koprulu M, Zengini E, Steinberg J, et al: Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 51:230–236. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Cheung KS, Sposito N, Stumpf PS, Wilson DI, Sanchez-Elsner T and Oreffo RO: MicroRNA-146a regulates human foetal femur derived skeletal stem cell differentiation by down-regulating SMAD2 and SMAD3. PLoS One. 9:e980632014. View Article : Google Scholar : PubMed/NCBI | |
|
Tardif G, Pelletier JP, Fahmi H, Hum D, Zhang Y, Kapoor M and Martel-Pelletier J: NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther. 15:R1972013. View Article : Google Scholar | |
|
Nishimura R, Hata K, Nakamura E, Murakami T and Takahata Y: Transcriptional network systems in cartilage development and disease. Histochem Cell Biol. 149:353–363. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Kanaan RA and Kanaan LA: Transforming growth factor beta1, bone connection. Med Sci Monit. 12:RA164–RA169. 2006.PubMed/NCBI | |
|
Dai J, Yu D, Wang Y, Chen Y, Sun H, Zhang X, Zhu S, Pan Z, Heng BC, Zhang S and Ouyang H: Kdm6b regulates cartilage development and homeostasis through anabolic metabolism. Ann Rheum Dis. 76:1295–1303. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Simon TC and Jeffries MA: The epigenomic landscape in osteoarthritis. Curr Rheumatol Rep. 19:302017. View Article : Google Scholar : PubMed/NCBI | |
|
Luo C, Hajkova P and Ecker JR: Dynamic DNA methylation: In the right place at the right time. Science. 361:1336–1340. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Barter MJ, Bui C, Cheung K, Falk J, Gómez R, Skelton AJ, Elliott HR, Reynard LN and Young DA: DNA hypomethylation during MSC chondrogenesis occurs predominantly at enhancer regions. Sci Rep. 10:11692020. View Article : Google Scholar : PubMed/NCBI | |
|
Takahashi A, de Andrés MC, Hashimoto K, Itoi E and Oreffo RO: Epigenetic regulation of interleukin-8, an inflammatory chemokine, in osteoarthritis. Osteoarthritis Cartilage. 23:1946–1954. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Shen J, Wang C, Li D, Xu T, Myers J, Ashton JM, Wang T, Zuscik MJ, McAlinden A and O'Keefe RJ: DNA methyltransferase 3b regulates articular cartilage homeostasis by altering metabolism. JCI Insight. 2:e936122017. View Article : Google Scholar : PubMed/NCBI | |
|
Hashimoto K, Oreffo RO, Gibson MB, Goldring MB and Roach HI: DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 60:3303–3313. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, Kokubun S and Bronner F: Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 52:3110–3124. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Goldring SR and Goldring MB: The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 427(427 Suppl): S27–S36. 2004. View Article : Google Scholar | |
|
Aida Y, Maeno M, Suzuki N, Namba A, Motohashi M, Matsumoto M, Makimura M and Matsumura H: The effect of IL-1beta on the expression of inflammatory cytokines and their receptors in human chondrocytes. Life Sci. 79:764–771. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Seto E and Yoshida M: Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 6:a0187132014. View Article : Google Scholar : PubMed/NCBI | |
|
Hong S, Derfoul A, Pereira-Mouries L and Hall DJ: A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. FASEB J. 23:3539–3552. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Huber LC, Brock M, Hemmatazad H, Giger OT, Moritz F, Trenkmann M, Distler JH, Gay RE, Kolling C, Moch H, et al: Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum. 56:1087–1093. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Meng F, Li Z, Zhang Z, Yang Z, Kang Y, Zhao X, Long D, Hu S, Gu M, He S, et al: MicroRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics. 8:2862–2883. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Cao K, Wei L, Zhang Z, Guo L, Zhang C, Li Y, Sun C, Sun X, Wang S, Li P and Wei X: Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: A novel mechanism of human osteoarthritis cartilage degeneration. Arthritis Res Ther. 16:4912014. View Article : Google Scholar | |
|
Higashiyama R, Miyaki S, Yamashita S, Yoshitaka T, Lindman G, Ito Y, Sasho T, Takahashi K, Lotz M and Asahara H: Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod Rheumatol. 20:11–17. 2010. View Article : Google Scholar : | |
|
Liao W, Sun J, Liu W, Li W, Jia J, Ou F, Su K, Zheng Y, Zhang Z and Sun Y: HDAC10 upregulation contributes to interleukin 1β-mediated inflammatory activation of synovium-derived mesenchymal stem cells in temporomandibular joint. J Cell Physiol. 234:12646–12662. 2019. View Article : Google Scholar | |
|
Jung JW, Lee S, Seo MS, Park SB, Kurtz A, Kang SK and Kang KS: Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol Life Sci. 67:1165–1176. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Dvir-Ginzberg M, Gagarina V, Lee EJ and Hall DJ: Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 283:36300–36310. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Tsuda M, Takahashi S, Takahashi Y and Asahara H: Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 278:27224–27229. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Fujita N, Matsushita T, Ishida K, Kubo S, Matsumoto T, Takayama K, Kurosaka M and Kuroda R: Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. J Orthop Res. 29:511–515. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Chen H, Liu X, Zhu W, Chen H, Hu X, Jiang Z, Xu Y, Wang L, Zhou Y, Chen P, et al: SIRT1 ameliorates age-related senescence of mesenchymal stem cells via modulating telomere shelterin. Front Aging Neurosci. 6:1032014. View Article : Google Scholar : PubMed/NCBI | |
|
Diao Z, Ji Q, Wu Z, Zhang W, Cai Y, Wang Z, Hu J, Liu Z, Wang Q, Bi S, et al: SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 49:4203–4219. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Fu Y, Kinter M, Hudson J, Humphries KM, Lane RS, White JR, Hakim M, Pan Y, Verdin E and Griffin TM: Aging promotes sirtuin 3-dependent cartilage superoxide dismutase 2 acetylation and osteoarthritis. Arthritis Rheumatol. 68:1887–1898. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wu Y, Chen L, Wang Y, Li W, Lin Y, Yu D, Zhang L, Li F and Pan Z: Overexpression of Sirtuin 6 suppresses cellular senescence and NF-κB mediated inflammatory responses in osteoarthritis development. Sci Rep. 5:176022015. View Article : Google Scholar | |
|
Collins JA, Kim CJ, Coleman A, Little A, Perez MM, Clarke EJ, Diekman B, Peffers MJ, Chubinskaya S, Tomlinson RE, et al: Cartilage-specific Sirt6 deficiency represses IGF-1 and enhances osteoarthritis severity in mice. Ann Rheum Dis. ard-2023-2243852023.Epub ahead of print. | |
|
Zhai XY, Yan P, Zhang J, Song HF, Yin WJ, Gong H, Li H, Wu J, Xie J and Li RK: Knockdown of SIRT6 enables human bone marrow mesenchymal stem cell senescence. Rejuvenation Res. 19:373–384. 2016. View Article : Google Scholar | |
|
Ji ML, Jiang H, Li Z, Geng R, Hu JZ, Lin YC and Lu J: Sirt6 attenuates chondrocyte senescence and osteoarthritis progression. Nat Commun. 13:76582022. View Article : Google Scholar : PubMed/NCBI | |
|
Wu SY, Du YC and Yue CF: Sirt7 protects chondrocytes degeneration in osteoarthritis via autophagy activation. Eur Rev Med Pharmacol Sci. 24:9246–9255. 2020.PubMed/NCBI | |
|
Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM and Chen D: Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 347:1374–1377. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Hsu YC, Wu YT, Tsai CL and Wei YH: Current understanding and future perspectives of the roles of sirtuins in the reprogramming and differentiation of pluripotent stem cells. Exp Biol Med (Maywood). 243:563–575. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Bi S, Liu Z, Wu Z, Wang Z, Liu X, Wang S, Ren J, Yao Y, Zhang W, Song M, et al: SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 11:483–504. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Hong S, Cho YW, Yu LR, Yu H, Veenstra TD and Ge K: Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 104:18439–18444. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ukita M, Matsushita K, Tamura M and Yamaguchi T: Histone H3K9 methylation is involved in temporomandibular joint osteoarthritis. Int J Mol Med. 45:607–614. 2020.PubMed/NCBI | |
|
Zhang F, Xu L, Xu L, Xu Q, Li D, Yang Y, Karsenty G and Chen CD: JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J Mol Cell Biol. 7:23–34. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Wang P, Li Y, Meng T, Zhang J, Wei Y, Meng Z, Lin Y, Liu D and Sui L: KDM6A promotes chondrogenic differentiation of periodontal ligament stem cells by demethylation of SOX9. Cell Prolif. 51:e124132018. View Article : Google Scholar | |
|
Lee HL, Yu B, Deng P, Wang CY and Hong C: Transforming growth factor-β-induced KDM4B promotes chondrogenic differentiation of human mesenchymal stem cells. Stem Cells. 34:711–719. 2016. View Article : Google Scholar | |
|
Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH and Wang CY: Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 11:50–61. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
He Q, Shi J, Liu W, Zhao W, Wang Z, Liu K, Zhao D, Wang S, Guo Y, Cheng L and Gao Y: TGF-β1-induced bone marrow mesenchymal stem cells (BMSCs) migration via histone demethylase KDM6B mediated inhibition of methylation marker H3K27me3. Cell Death Discov. 8:3392022. View Article : Google Scholar | |
|
Duan L, Liang Y, Xu X, Wang J, Li X, Sun D, Deng Z, Li W and Wang D: Noncoding RNAs in subchondral bone osteoclast function and their therapeutic potential for osteoarthritis. Arthritis Res Ther. 22:2792020. View Article : Google Scholar : PubMed/NCBI | |
|
van Meurs JB, Boer CG, Lopez-Delgado L and Riancho JA: Role of epigenomics in bone and cartilage disease. J Bone Miner Res. 34:215–230. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Nakamura Y, Inloes JB, Katagiri T and Kobayashi T: Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 31:3019–3028. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al: MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24:1173–1185. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Iliopoulos D, Malizos KN, Oikonomou P and Tsezou A: Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 3:e37402008. View Article : Google Scholar : PubMed/NCBI | |
|
Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK and Asahara H: MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 60:2723–2730. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Ji Q, Xu X, Zhang Q, Kang L, Xu Y, Zhang K, Li L, Liang Y, Hong T, Ye Q and Wang Y: The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis. J Mol Med (Berl). 94:771–785. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Mao G, Zhang Z, Huang Z, Chen W, Huang G, Meng F, Zhang Z and Kang Y: MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthritis Cartilage. 25:521–532. 2017. View Article : Google Scholar | |
|
Mao G, Wu P, Zhang Z, Zhang Z, Liao W, Li Y and Kang Y: MicroRNA-92a-3p regulates aggrecanase-1 and aggrecanase-2 expression in chondrogenesis and IL-1β-induced catabolism in human articular chondrocytes. Cell Physiol Biochem. 44:38–52. 2017. View Article : Google Scholar | |
|
Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR and Haqqi TM: MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 62:1361–1371. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Yang B, Guo H, Zhang Y, Chen L, Ying D and Dong S: MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 6:e216792011. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Gibson G, Kim JS, Kroin J, Xu S, van Wijnen AJ and Im HJ: MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 480:34–41. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Vonk LA, Kragten AH, Dhert WJ, Saris DB and Creemers LB: Overexpression of hsa-miR-148a promotes cartilage production and inhibits cartilage degradation by osteoarthritic chondrocytes. Osteoarthritis Cartilage. 22:145–153. 2014. View Article : Google Scholar | |
|
Joung S, Yoon DS, Cho S, Ko EA, Lee KM, Park KH, Lee JW and Kim SH: Downregulation of MicroRNA-495 alleviates IL-1β responses among chondrocytes by preventing SOX9 reduction. Yonsei Med J. 62:650–659. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Qiu M, Liu D and Fu Q: MiR-129-5p shuttled by human synovial mesenchymal stem cell-derived exosomes relieves IL-1β induced osteoarthritis via targeting HMGB1. Life Sci. 269:1189872021. View Article : Google Scholar | |
|
Zhou JX, Tian ZG, Zhu LF, Wu WD, Zhou SL, Zhao YT and Huang S: MicroRNA-615-3p promotes the osteoarthritis progression by inhibiting chondrogenic differentiation of bone marrow mesenchymal stem cells. Eur Rev Med Pharmacol Sci. 22:6212–6220. 2018.PubMed/NCBI | |
|
Lv S, Xu J, Chen L, Wu H, Feng W, Zheng Y, Li P, Zhang H, Zhang L, Chi G and Li Y: MicroRNA-27b targets CBFB to inhibit differentiation of human bone marrow mesenchymal stem cells into hypertrophic chondrocytes. Stem Cell Res Ther. 11:3922020. View Article : Google Scholar : PubMed/NCBI | |
|
Huang T, Zhou Y, Wang J, Cao Y and Hang DH: MiR-26b regulates cartilage differentiation of bone marrow mesenchymal stem cells in rats through the Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 23:5084–5092. 2019.PubMed/NCBI | |
|
Chen HO, Zhang L, Tang ZY and Gong ZM: MiR-485-5p promotes the development of osteoarthritis by inhibiting cartilage differentiation in BMSCs. Eur Rev Med Pharmacol Sci. 22:3294–3302. 2018.PubMed/NCBI | |
|
Qin F, Wang F, Wang XP, Chen J, Zeng FH, Sun CL, Mao JP and Li CL: MiR-539-3p inhibited chondrogenic differentiation in human adipose stem cells by targeting Sox9. J Orthop Surg Res. 17:1682022. View Article : Google Scholar : PubMed/NCBI | |
|
Yang M, Yan X, Yuan FZ, Ye J, Du MZ, Mao ZM, Xu BB, Chen YR, Song YF, Fan BS and Yu JK: MicroRNA-210-3p promotes chondrogenic differentiation and inhibits adipogenic differentiation correlated with HIF-3α signalling in bone marrow mesenchymal stem cells. Biomed Res Int. 2021:66999102021. | |
|
Zhang P, Gao G, Zhou Z and He X: microRNA-130b downregulation potentiates chondrogenic differentiation of bone marrow mesenchymal stem cells by targeting SOX9. Braz J Med Biol Res. 54:e103452021. View Article : Google Scholar : PubMed/NCBI | |
|
Feng L, Yang ZM, Li YC, Wang HX, Lo JHT, Zhang XT and Li G: Linc-ROR promotes mesenchymal stem cells chondrogenesis and cartilage formation via regulating SOX9 expression. Osteoarthritis Cartilage. 29:568–578. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Alahdal M, Huang R, Duan L, Zhiqin D, Hongwei O, Li W and Wang D: Indoleamine 2, 3 dioxygenase 1 impairs chondrogenic differentiation of mesenchymal stem cells in the joint of osteoarthritis mice model. Front Immunol. 12:7811852021. View Article : Google Scholar : PubMed/NCBI | |
|
Ma L, Bajic VB and Zhang Z: On the classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ou F, Su K, Sun J, Liao W, Yao Y, Zheng Y and Zhang Z: The LncRNA ZBED3-AS1 induces chondrogenesis of human synovial fluid mesenchymal stem cells. Biochem Biophys Res Commun. 487:457–463. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Huynh NP, Gloss CC, Lorentz J, Tang R, Brunger JM, McAlinden A, Zhang B and Guilak F: Long non-coding RNA GRASLND enhances chondrogenesis via suppression of the interferon type II signaling pathway. Elife. 9:e495582020. View Article : Google Scholar : PubMed/NCBI | |
|
Fang P, Zhang LX, Hu Y, Zhang L and Zhou LW: Long non-coding RNA DANCR induces chondrogenesis by regulating the miR-1275/MMP-13 axis in synovial fluid-derived mesenchymal stem cells. Eur Rev Med Pharmacol Sci. 23:10459–10469. 2019.PubMed/NCBI | |
|
Ji Y, Fang QY, Wang SN, Zhang ZW, Hou ZJ, Li JN and Fu SQ: Lnc-RNA BLACAT1 regulates differentiation of bone marrow stromal stem cells by targeting miR-142-5p in osteoarthritis. Eur Rev Med Pharmacol Sci. 24:2893–2901. 2020.PubMed/NCBI | |
|
Zhu Y, Li R and Wen LM: Long non-coding RNA XIST regulates chondrogenic differentiation of synovium-derived mesenchymal stem cells from temporomandibular joint via miR-27b-3p/ADAMTS-5 axis. Cytokine. 137:1553522021. View Article : Google Scholar | |
|
Chen H, Yang S and Shao R: Long non-coding XIST raises methylation of TIMP-3 promoter to regulate collagen degradation in osteoarthritic chondrocytes after tibial plateau fracture. Arthritis Res Ther. 21:2712019. View Article : Google Scholar : PubMed/NCBI | |
|
Wen C, Lin L, Zou R, Lin F and Liu Y: Mesenchymal stem cell-derived exosome mediated long non-coding RNA KLF3-AS1 represses autophagy and apoptosis of chondrocytes in osteoarthritis. Cell Cycle. 21:289–303. 2022. View Article : Google Scholar : | |
|
Liu Y, Zou R, Wang Z, Wen C, Zhang F and Lin F: Exosomal KLF3-AS1 from hMSCs promoted cartilage repair and chondrocyte proliferation in osteoarthritis. Biochem J. 475:3629–3638. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Li J, Shao W and Shen N: LncRNA CTBP1-AS2 is upregulated in osteoarthritis and increases the methylation of miR-130a gene to inhibit chondrocyte proliferation. Clin Rheumatol. 39:3473–3478. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu J, Fu Q, Shao J, Peng J, Qian Q, Zhou Y and Chen Y: Regulating effect of Circ_ATRNL1 on the promotion of SOX9 expression to promote chondrogenic differentiation of hAMSCs mediated by MiR-145-5p. J Tissue Eng Regen Med. 15:487–502. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Li S, Liu J, Liu S, Jiao W and Wang X: Mesenchymal stem cell-derived extracellular vesicles prevent the development of osteoarthritis via the circHIPK3/miR-124-3p/MYH9 axis. J Nanobiotechnology. 19:1942021. View Article : Google Scholar : PubMed/NCBI | |
|
Bao C and He C: The role and therapeutic potential of MSC-derived exosomes in osteoarthritis. Arch Biochem Biophys. 710:1090022021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Lin L, Zou R, Wen C, Wang Z and Lin F: MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 17:2411–2422. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Mao G, Zhang Z, Hu S, Zhang Z, Chang Z, Huang Z, Liao W and Kang Y: Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res Ther. 9:2472018. View Article : Google Scholar : PubMed/NCBI | |
|
Tao Y, Zhou J, Wang Z, Tao H, Bai J, Ge G, Li W, Zhang W, Hao Y, Yang X and Geng D: Human bone mesenchymal stem cells-derived exosomal miRNA-361-5p alleviates osteoarthritis by downregulating DDX20 and inactivating the NF-κB signaling pathway. Bioorg Chem. 113:1049782021. View Article : Google Scholar | |
|
Cosenza S, Ruiz M, Toupet K, Jorgensen C and Noël D: Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci Rep. 7:162142017. View Article : Google Scholar : PubMed/NCBI | |
|
Xia Q, Wang Q, Lin F and Wang J: miR-125a-5p-abundant exosomes derived from mesenchymal stem cells suppress chondrocyte degeneration via targeting E2F2 in traumatic osteoarthritis. Bioengineered. 12:11225–11238. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Chen X, Shi Y, Xue P, Ma X, Li J and Zhang J: Mesenchymal stem cell-derived exosomal microRNA-136-5p inhibits chondrocyte degeneration in traumatic osteoarthritis by targeting ELF3. Arthritis Res Ther. 22:2562020. View Article : Google Scholar : PubMed/NCBI | |
|
Woo CH, Kim HK, Jung GY, Jung YJ, Lee KS, Yun YE, Han J, Lee J, Kim WS, Choi JS, et al: Small extracellular vesicles from human adipose-derived stem cells attenuate cartilage degeneration. J Extracell Vesicles. 9:17352492020. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Z, Yan K, Ge G, Zhang D, Bai J, Guo X, Zhou J, Xu T, Xu M, Long X, et al: Exosomes derived from miR-155-5p-overexpressing synovial mesenchymal stem cells prevent osteoarthritis via enhancing proliferation and migration, attenuating apoptosis, and modulating extracellular matrix secretion in chondrocytes. Cell Biol Toxicol. 37:85–96. 2021. View Article : Google Scholar | |
|
Lu L, Wang J, Fan A, Wang P, Chen R, Lu L and Yin F: Synovial mesenchymal stem cell-derived extracellular vesicles containing microRN555A-26a-5p ameliorate cartilage damage of osteoarthritis. J Gene Med. 23:e33792021. View Article : Google Scholar : PubMed/NCBI | |
|
Zeng Z, Dai Y, Deng S, Zou S, Dou T and Wei F: Synovial mesenchymal stem cell-derived extracellular vesicles alleviate chondrocyte damage during osteoarthritis through microRNA-130b-3p-mediated inhibition of the LRP12/AKT/β-catenin axis. Immunopharmacol Immunotoxicol. 44:247–260. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Kong R, Gao J, Zhang J, Ji L, Yu Y, Zhang L and Zhao D: Synovial mesenchymal stem cell-derived exosomal miR-320c enhances chondrogenesis by targeting ADAM19. Future Med Chem. 14:81–96. 2022. View Article : Google Scholar | |
|
Liu X, Liu Y, He H, Xiang W and He C: Human adipose and synovial mesenchymal stem cells improve osteoarthritis in rats by reducing chondrocyte reactive oxygen species and inhibiting inflammatory response. J Clin Lab Anal. 36:e243532022. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Yu D, Liu Z, Zhou F, Dai J, Wu B, Zhou J, Heng BC, Zou XH, Ouyang H and Liu H: Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther. 8:1892017. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Zeng Y, Si HB, Tang L, Xie HQ and Shen B: Exosomes derived from human urine-derived stem cells overexpressing miR-140-5p alleviate knee osteoarthritis through downregulation of VEGFA in a rat model. Am J Sports Med. 50:1088–1105. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Mao G, Hu S, Zhang Z, Wu P, Zhao X, Lin R, Liao W and Kang Y: Exosomal miR-95-5p regulates chondrogenesis and cartilage degradation via histone deacetylase 2/8. J Cell Mol Med. 22:5354–5366. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Shema E, Bernstein BE and Buenrostro JD: Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat Genet. 51:19–25. 2019. View Article : Google Scholar | |
|
Chan CKF, Gulati GS, Sinha R, Tompkins JV, Lopez M, Carter AC, Ransom RC, Reinisch A, Wearda T, Murphy M, et al: Identification of the human skeletal stem cell. Cell. 175:43–56.e21. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al: Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 119:555–566. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Ji Q, Zheng Y, Zhang G, Hu Y, Fan X, Hou Y, Wen L, Li L, Xu Y, Wang Y and Tang F: Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 78:100–110. 2019. View Article : Google Scholar | |
|
Hu X, Li Z, Ji M, Lin Y, Chen Y and Lu J: Identification of cellular heterogeneity and immunogenicity of chondrocytes via single-cell RNA sequencing technique in human osteoarthritis. Front Pharmacol. 13:10047662022. View Article : Google Scholar : PubMed/NCBI | |
|
Chou CH, Jain V, Gibson J, Attarian DE, Haraden CA, Yohn CB, Laberge RM, Gregory S and Kraus VB: Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep. 10:108682020. View Article : Google Scholar : PubMed/NCBI | |
|
Sebastian A, McCool JL, Hum NR, Murugesh DK, Wilson SP, Christiansen BA and Loots GG: Single-cell RNA-Seq reveals transcriptomic heterogeneity and post-traumatic osteoarthritis-associated early molecular changes in mouse articular chondrocytes. Cells. 10:14622021. View Article : Google Scholar : PubMed/NCBI | |
|
Pengas I, Eldridge S, Assiotis A, McNicholas M, Mendes JE and Laver L: MMP-3 in the peripheral serum as a biomarker of knee osteoarthritis, 40 years after open total knee meniscectomy. J Exp Ortho. 5:212018. View Article : Google Scholar | |
|
Chou CH, Lee MT, Song IW, Lu LS, Shen HC, Lee CH, Wu JY, Chen YT, Kraus VB and Wu CC: Insights into osteoarthritis progression revealed by analyses of both knee tibiofemoral compartments. Osteoarthritis Cartilage. 23:571–580. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Qu Y, Wang Y, Wang S, Yu X, He Y, Lu R, Chen S, Meng C, Xu H, Pei W, et al: A comprehensive analysis of single-cell RNA transcriptome reveals unique SPP1+ chondrocytes in human osteoarthritis. Comput Biol Med. 160:1069262023. View Article : Google Scholar : PubMed/NCBI | |
|
Gao C, Pu H, Zhou Q, Tao T, Liu H, Sun X, He X and Xiao J: Two reactive behaviors of chondrocytes in an IL-1β-induced inflammatory environment revealed by the single-cell RNA sequencing. Aging (Albany NY). 13:11646–11664. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Yoshimoto M, Sadamori K, Tokumura K, Tanaka Y, Fukasawa K and Hinoi E: Bioinformatic analysis reveals potential relationship between chondrocyte senescence and protein glycosylation in osteoarthritis pathogenesis. Front Endocrinol (Lausanne). 14:11536892023. View Article : Google Scholar : PubMed/NCBI |