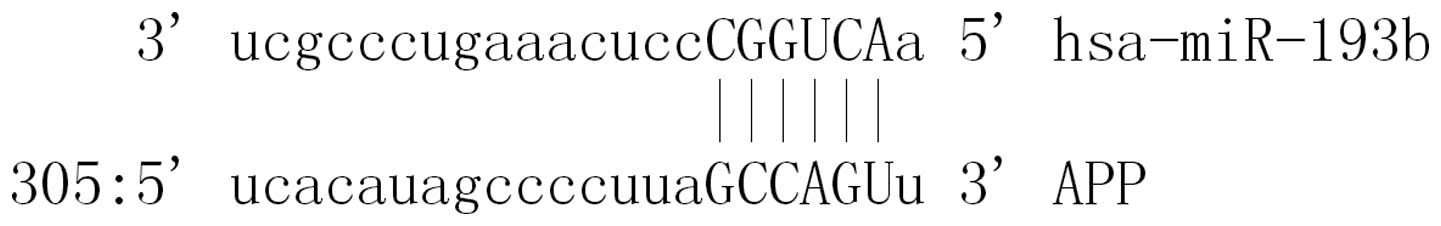MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer's disease
- Authors:
- Published online on: August 12, 2014 https://doi.org/10.3892/mmr.2014.2484
- Pages: 2395-2400
Abstract
Introduction
Alzheimer’s disease (AD) is a prominent neurodegenerative disorder characterized by progressive loss of memory and other cognitive functions. Despite considerable progress in genetics and cell biology, numerous questions remain regarding the mechanisms of neurodegeneration and the molecular and pathological components. Extracellular amyloid-β (Aβ), which is derived from the larger amyloid precursor protein (APP), is considered to be responsible for the death of neurons and dementia in Alzheimer’s disease. An increased expression of APP may increase the risk of AD (1,2). APP levels can be regulated at the genomic, transcriptional or translational level, and in the rate of degradation. Genetic variants in the APP promoter increase APP transcription by 2–3 fold and have been reported to increase the risk of AD (2). APP can be processed by a group of secretases where α-secretase produces soluble fragments, whereas β- and γ-secretase generate Aβ from APP (3). Previous research has suggested that Aβ regulates neuronal and synaptic activity, and accumulation of Aβ in the brain causes a combination of aberrant network activity and synaptic depression (4).
MicroRNAs (miRs) are endogenous, short, noncoding RNAs. Mature miRs are single-stranded RNA molecules of ~20–25 nucleotides which act as important post-transcriptional regulators of gene expression by binding with their target mRNAs. They are additionally essential for normal neuronal function and survival (3,4). Several miRs have been shown to be important in neuropathology by downregulating AD-related proteins, including APP and BACE-1. It has been demonstrated that miR-16, -101, -106a/b, -147 and -160a could function as APP suppressors (5–8).
Several cerebral spinal fluid (CSF) or blood-based markers, such as Aβ, tau and phosphorylated tau (p-tau) are proposed biomarkers for predicting future cognitive decline in healthy individuals, and the progression to dementia in patients who are cognitively impaired (9–11). Previous studies have shown that APP and BACE-1 are novel markers of AD (12–14). However, there is still an urgent requirement for additional biomarkers that can detect AD in the pre-dementia phase (9–14). Alterations to the regulation of miRs in the blood and CSF may indicate progression in AD (15,16). Exosomes are membrane vesicles with a size of 40–100 nm that are released from numerous cell types of the body (17). Recent studies have shown that, in addition to functional proteins, exosomes carry mRNA as well as miRs (18). In functional terms, exosomes are considered to represent a novel mechanism of intercellular communication. This may be due to the uptake of exosomes by target cells or by triggering cell signaling through membrane receptors (17–19). In a previous study, we found that miR-193b was decreased in the hippocampi of 9-month-old APP/PS1 double-transgenic mice by miR array. In the present study, bioinformatic analyses showed that miR-193b may potentially target the 3′-untranslated region (UTR) of APP and the effects of miR-193b on the expression of APP were studied. Blood and CSF derived exosomal miR-193b was detected in APP/PS1 double-transgenic mice, and patients with mild cognitive impairment (MCI) and dementia of Alzheimer type (DAT).
Materials and methods
Study population
The design of the present study was approved by the ethics committee of Xuanwu Hospital of Capital Medical University (Beijing, China), and the written informed consents were obtained from all participants. A total of 43 MCI (23 females, 20 males, mean age 63.8±6.1) and 51 DAT patients (28 females, 23 males, mean age 64.2±6.5) were selected for this study. The CSF was drawn within 2 h following collection of blood (n=7). Age- and gender-matched control subjects were included in the experimental design. Samples were stored at −80°C until required for further analysis. The expression levels of Aβ, tau and p-tau in the plasma and CSF of the subjects were determined by ELISA kit from Cusabio (Wuhan, China). The study was approved by the Ethics Committee of Xuanwu Hospital of Capital Medical University (Beijing, China). Written informed consent was obtained from the patients’ family.
APP/PS1 double-transgenic and wild-type mice
The 3, 6 and 9-month-old APP/PS1 double-transgenic mice on a C57BL/6J genetic background were purchased from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences & Comparative Medical Center (Beijing, China). All the animal protocols were approved by Ethics Committee of Xuanwu Hospital of Capital Medical University (Beijing, China). The non-transgenic mice were used as wild-type (WT) controls. Blood was taken by removing the eyeballs and CSF-like fluid was collected as previously described (20). The hippocampi were isolated as described for miR-193b quantitative polymerase chain reaction (qPCR) detection (n=5). The samples were stored in liquid nitrogen until analysis.
Cell culture and miR transfections
SH-SY5Y and HEK293 cell lines were purchased from Shanghai Institute of Cell Biology, China. Cells were grown in antibiotic-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. SH-SY5Y cells were transfected with 100 nM (final concentration) miR-193b mimic oligonucleotide, miR-193b inhibitor oligonucleotide or a non-specific control small interfering RNA siRNA) (GenePharma) using Lipofectamine™ 2000 reagent (Invitrogen Life Sciences, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Reporter vectors and DNA constructs
Reporter vectors containing the putative miR-193b target site from the APP 3′-UTR was synthesized with double-stranded oligos perfectly complementary to putative miR-193b target site and oligos in which the seed regions were mutated (21). The APP oligos had the sequence (seed region bolded) as follows: 5′-CCCAAGCTTTCACATA GCCCCTTAGCCATTTAAGCTTGGG-3′; 3′-GGGT TCGAAAGTGTATCGGGGAATCGGTAAATTCGAACC-5′. The mutant APP target oligos had nucleotides 3–6 of the seed region mutated (italicized): 5′-CCCAAGCTTTCAC ATAGCCCCTTAACTAGTTAAGCTTGGG-3′; 3′-GGGTTC GAAAGTGTATCGGGGAATTGATCAATTCGAACCC-5′. HEK-293 cells were plated in 24-well plates. The following day, cells were transfected with a miR mimic oligonucleotide, reporter vectors bearing either the miR target sequence or the miR seed region mutant target sequence, and one tenth of the molar volume of pRL-SV40, a Renilla luciferase control vector. Arrest-In transfection reagent (Open Biosystems Inc; GE healthcare, Fairfield, CT, USA) was used; any differences in transfection efficiency were accounted for by measuring Renilla luciferase activity. Following 48 h posttransfection, cells were lysed using 100 μl of GLB (Glo Lysis Buffer, Promega Corporation, Madison, WI, USA). Firefly and Renilla luciferase activities were measured using a dual luciferase reporter assay kit (Promega), according to the manufacturer’s instructions. Firefly luciferase activity was normalized to the Renilla luciferase activity.
Isolation of RNA and APP mRNA qPCR analysis
Total RNA from harvested cells was isolated using TRIzol™ Reagent (Invitrogen Life Sciences) according to the manufacturer’s instructions. Isolated RNA was reverse transcribed using the PrimeScript™ RT reagent (Takara Bio, Inc., Shiga, Japan). The mRNA expressions of APP was determined using SYBR® Green qPCR (Takara Bio, Inc) using a LightCycler 480 System (Roche Diagnostics, Mannheim, Germany). GAPDH was used to normalize the target genes. The PCR primer sequences were as follows: APP, forward, 5′-TTGCGAAACTCATCTTCACTGG-3′, reverse 5′-CAGTGGGCAACACACAAACTCTAC-3′; GAPDH, forward, 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse 5′-TGGTGAAGACGCCAGTGGA-3′. The number of samples in each group was five.
Western blots
Western blotting was performed as previously described (22). Cells were homogenized in extraction buffer, then incubated at 0°C for 15 min and centrifuged at 2,000 × g for 10 min at 4°C. Aliquots of the supernatants were used for the measurement of the total protein content. Samples were boiled for 5 min in loading buffer and then the proteins (30 μg per well) were separated by 10% SDS-PAGE (Bio-Rad, Hercules, CA, USA). Proteins in the gel were transferred onto a nitrocellulose membrane (Pall Corporation, New York, NY, USA). Membranes were incubated with anti-APP (diluted 1:500; Abcam, Cambridge, UK), and anti-GAPDH (diluted 1:400, Abcam) at room temperature for 1.5 h, respectively. Membranes were then washed and incubated with anti-IgG antibody linked to horseradish peroxidase at room temperature for 1 h. The membranes were then incubated with substrate for peroxidase and enhanced chemiluminescence (KPL, Gaithersburg, MD, USA) for 1 min and exposed immediately to X-ray film for 1–5 min. Films were then revealed in the conventional manner. The amount of each protein was measured by densitometric analysis and standardized relative to GAPDH. The number of samples in each group was five.
Isolation of exosomes
The exosomes were isolated using the Total Exosome Isolation kit (Invitrogen Life Technologies) according to the manufacturer’s instructions. Briefly, the serum was centrifuged at 2,000 × g for 30 min to remove cells and debris. Following this, 400 μl clarified serum was transferred to a new tube and 0.4 volumes of the Total Exosome Isolation reagent was added. The serum/reagent solution was mixed and then incubated at 4°C for 30 min. After incubation, the samples were centrifuged at 10,000 × g for 10 min at room temperature. The supernatant was discarded and the pellet, containing the exosomes, at the bottom of the tube was resuspended in 200 μl phosphate-buffered saline (PBS).
Isolation of RNA and miR-193b qPCR analysis
Total RNA in 350 μl CSF, or 200 μl serum samples was extracted using a spin column method with an miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Exosomal RNA was isolated and purified using a Total Exosome RNA Isolation kit (Invitrogen Life Technologies). The RNA isolated from the CSF was 530–1,500 ng/ml, 2,100–4,700 ng/ml from serum, 270–420 ng/ml from the exosome. Total RNA in hippocampus tissue from the animal models and cultured cells was extracted using a spin column method using the miRNeasy kit (Qiagen). MiRs were reverse transcribed into cDNA using the miScript II RT kit (Qiagen) in a 10 μl reaction system. MiR-193b were determined by a TaqMan qPCR method (Applied Biosystems, Foster City, CA, USA), using U6 RNA as an endogenous control.
Statistical analyses
Statistical analyses were performed using SPSS 13.0 for Windows (SPSS, Inc., Chicago, IL, USA). For normally distributed data, results are expressed as the mean ± standard deviation. The differences between groups were assessed by t-tests and analyzed using the Mann-Whitney U-test. Correlations were determined by computing the Spearman rank correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
Bioinformatics retrieval
A total of 34 miRs were found to be putative targets on the 3′-UTR of APP. MiR-193b was a miR that may target the 3′-UTRs of APP (Fig. 1).
MiR-193b represses the mRNA and protein expression of APP
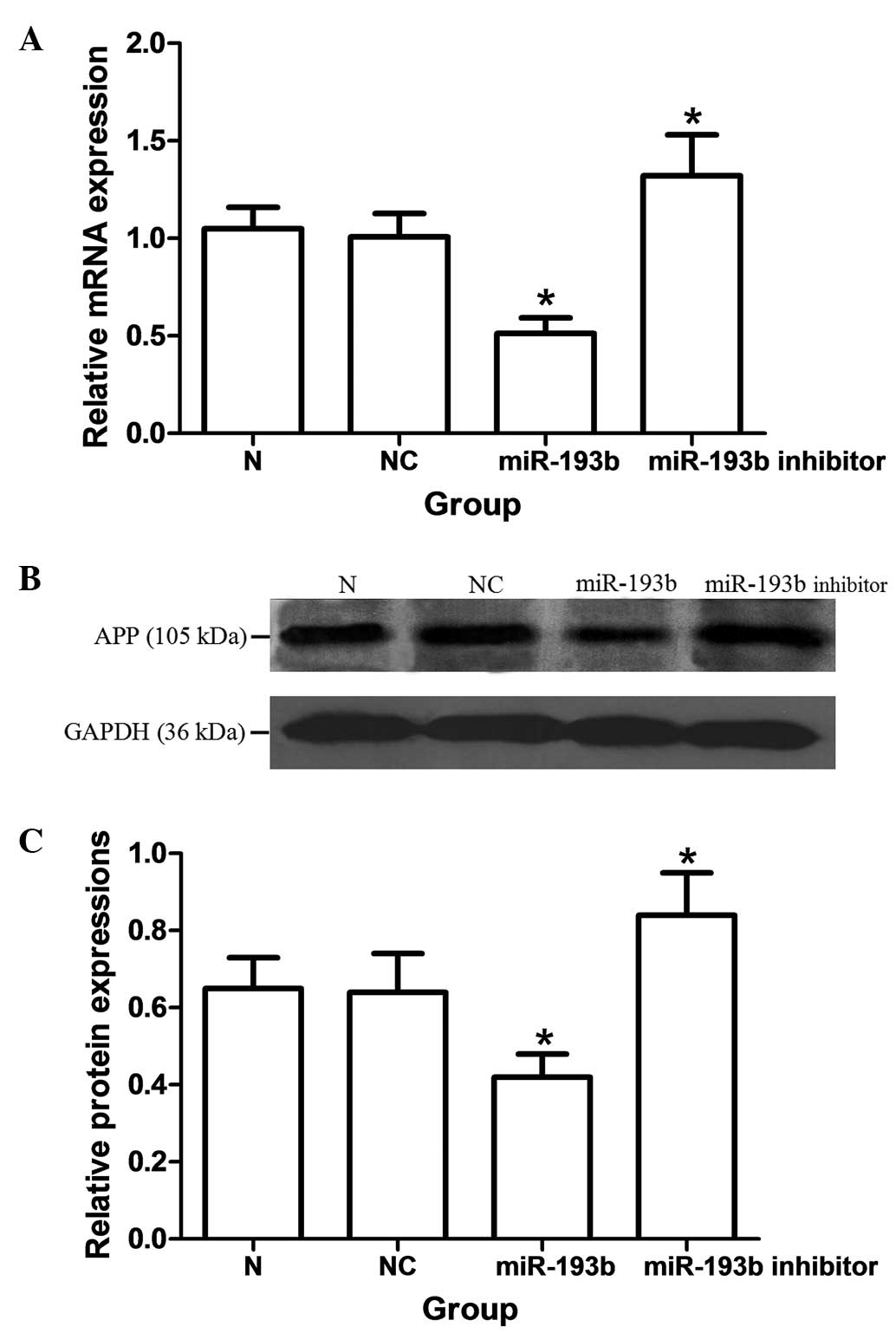
As shown in Fig. 2, the mRNA and protein expressions of APP were significantly decreased by miRNA-193b in SH-SY5Y cells (P<0.05). The miR-193b inhibitor oligonucleotide induced a significant upregulation of mRNA and protein expression of APP as compared with the nonspecific control groups, respectively (P<0.05). By TaqMan qPCR, a ~55% downregulation of endogenous miR-193b was observed (data not shown).
APP 3′-UTR is a miR-193b target
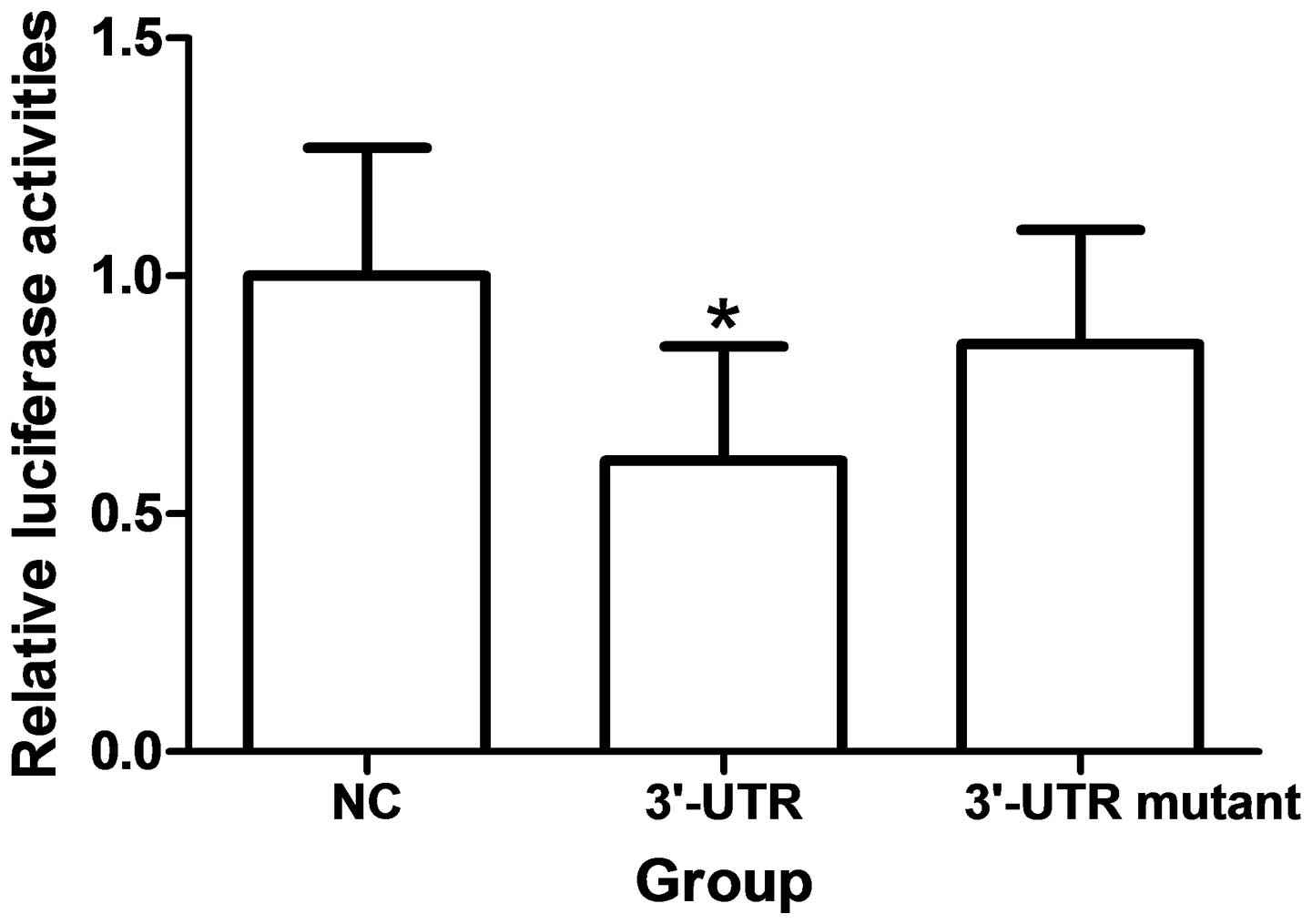
Over-expression of miR-193b significantly reduces fluorescence from APP reporter vectors in HEK293 cells (P<0.05). These reductions were not observed when mutations were made to the 3′UTR seed regions of APP or BACE-1 were generated (Fig. 3).
Exosomal miR-193b decreases in CSF-like fluid and serum of transgenic mice
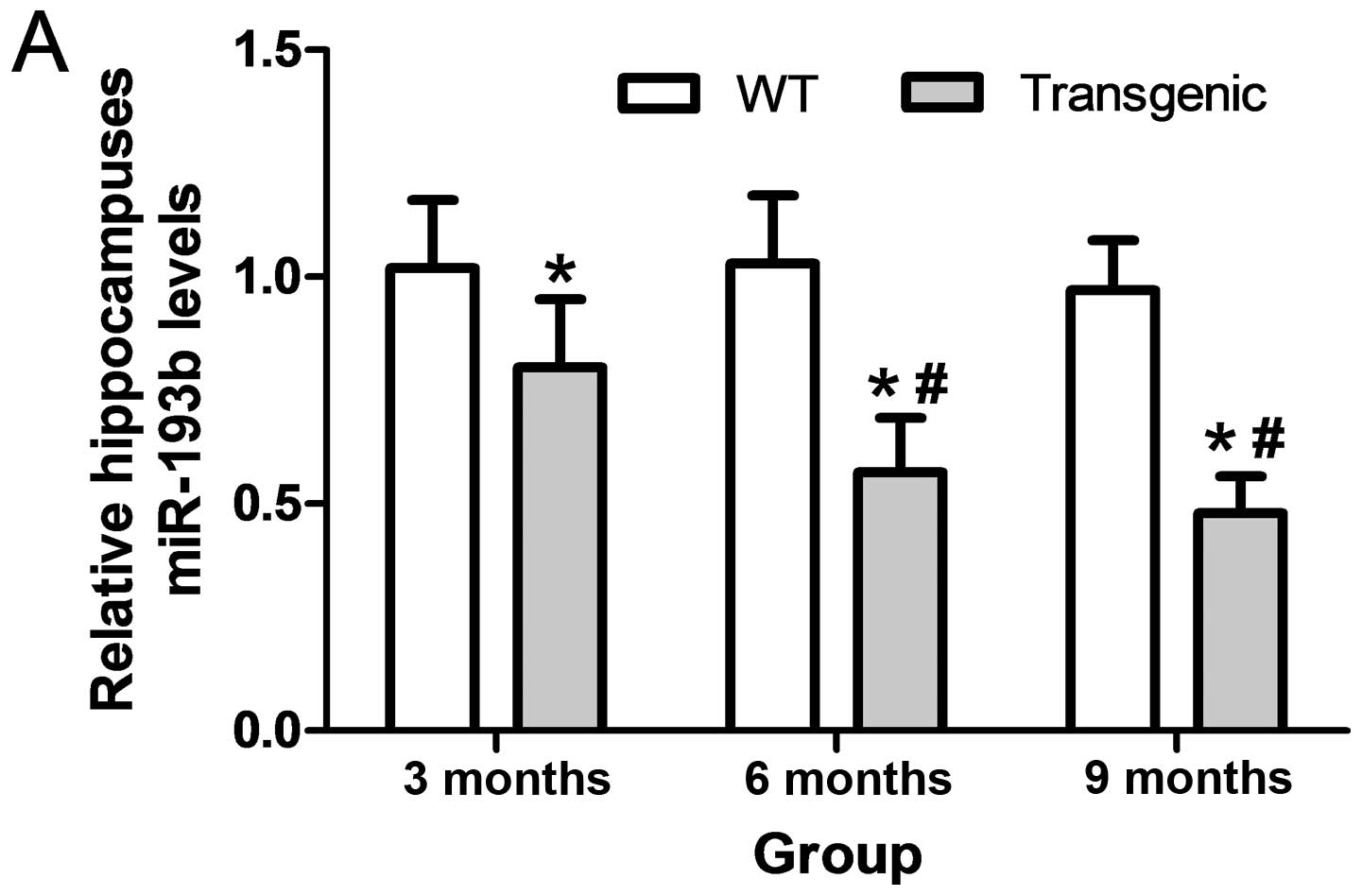
The levels of miR-193b were significantly downregulated in the hippocampi of 3, 6 and 9 month APP/PS1 transgenic mice as compared with the WT mice (P<0.05). The levels of exosomal miR-193b were significantly downregulated in the CSF-like fluid and the serum of 3, 6 and 9 month APP/PS1 transgenic mice, as compared with the WT mice (P<0.05). Levels of exosomal miR-193b in the CSF-like fluid and serum of 6 and 9 month transgenic mice were significantly lower than that of the 3 month transgenic mice, respectively (P<0.05; Fig. 4).
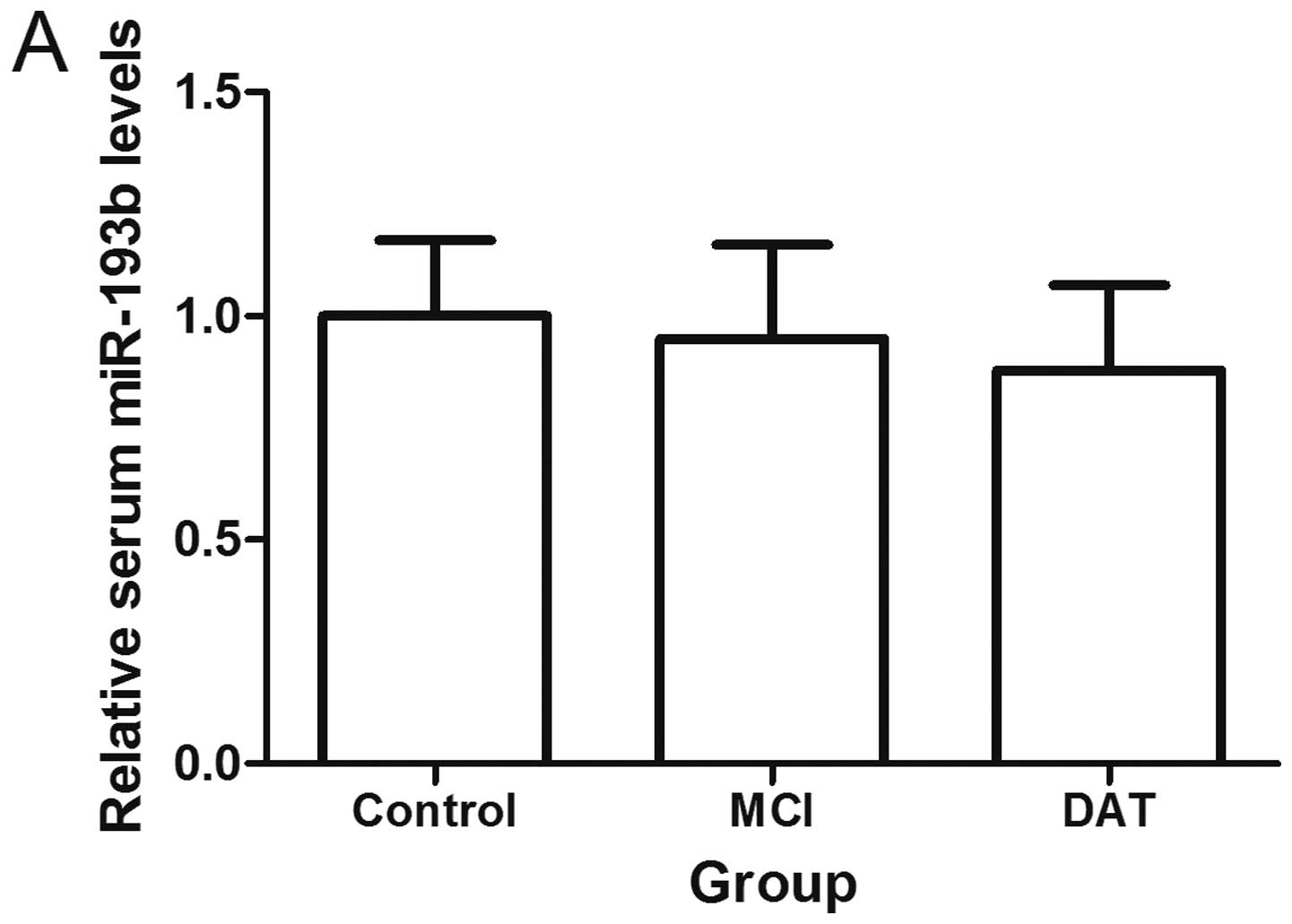
Exosomal miR-193b decreases in the CSF, serum and plasma of AD patients
Compared with the control groups, patients with MCI and DAT had lower levels of exosomal miR-193b in the serum and plasma (P<0.05). Patients with DAT had lower exosomal miR-193b levels in their serum and plasma compared with the MCI groups (P<0.05). It was additionally found that exosomal miR-193b was decreased in the CSF of patients with DAT as compared with the control group (n=7; P<0.05; Fig. 5).
The level of exosomal miR-193b was lower in the CSF compared with serum from a given individual (P<0.05; data not shown). There was no correlation between exosomal miRNA-193b levels and the CSF and serum from a given individual (data not shown). When the cutoff value was set as the mean concentration - two standard deviations of the controls, the positive rates of exosomal miR-193b were 71.43% (5/7) and 58.82% (30/51) in the CSF and serum of patients with DAT respectively, and 58.14% (25/43) in the serum of patients with MCI.
Exosomal miR-193b is negatively correlated with Aβ42 in the CSF but not in the serum
A weak, but significant, negative correlation was found between the levels of exosomal miR-193b and Aβ42 in the CSF of patients with DAT (r =−0.442, P<0.05), as well as the control group (r =−0.503, P<0.05). MiR-193b and exosomal miR-193b had no correlation with HCY, ApoE, tau and p-tau in the serum and CSF (data not shown).
Discussion
The potential benefit in the analysis of miR in the diagnosis and treatment of numerous diseases, including cancer, infection and neurodegenerative disease has been previously evaluated by numerous researchers (6,23,24). The expression profiles of miRs are known to be altered in several regions of the AD brain, however the cause or consequence in the pathology of the disease is unknown. There has been no data to suggest a direct genetic link between miRs or miR recognition elements and neurodegenerative disease (6,9,10,23,24). High expression of APP correlating with accelerated accumulation of Aβ in the brain is a feature of AD. Previous research has demonstrated miR-106a/b, -147, -160a, -520c and -655 function as APP suppressors (5–8). In the present study, it was shown that miR-193b could repress the expression of APP by binding its 3′-UTR and decay its mRNA. The deregulated miR-193b may play a role in the development of AD and the downregulating effects of miR-193b on APP may represent a study direction for AD therapy by miR.
Previous studies have demonstrated that miRs are stably expressed in various bodily fluids, and their unique expression patterns can serve as fingerprints of various diseases, including AD (25). The CSF is in direct contact with the extracellular space of the brain and can reflect biochemical changes that occur in the latter. The CSF is therefore the optimal source of AD biomarkers. CSF, however, is not an appropriate sample for screening and routine testing due to its invasive process of sample collection. From this perspective, blood based biomarkers for AD screening and routine testing would be more suitable (26). Circulating miRs can derive from many sources, including cell death and lysed cells, which are passive secretions, as well as active secretion from cells by exosomes and microvesicles. Although endogenous plasma miRNAs exist in a form that is resistant to plasma RNase activity and other conditions such as extreme pH, the exosomes may provide additional protection, which allows data to be obtained that reflects the accurate expression level of miR actively secreted from cells. The miRs can also be transported by other carriers such as high and low density lipoprotein, both of which are highly abundant in plasma and cannot be generated by brain (27). The total small RNAs isolated from body fluid may contain more non-brain-derived miRs than that from exosomes. Furthermore, exosomes are actively secreted from cells, which can help to eliminate the interference from passively secreted miRs.
APP/PS-1 double-transgenic mice contained insoluble amyloid peptides at the age of 6–9 months, concomitant with the formation of amyloid plaques (28). In the present study, it was shown that the level of miR-193b was decreased in the hippocampi of 3 month transgenic mice, which suggested that the change of miR-193b is earlier than the formation of amyloid plaques. The detection of exosomal miR-193b in the CSF-like fluid and plasma of 3, 6 and 9 month transgenic mice demonstrated that exosomal miR-193b was a potential AD biomarker, especially for the earlier stages of the disease. This conjecture was further confirmed by clinical detection, which showed that exosomal miR-193b levels in patients with MCI were higher as compared with the control group and lower as compared with the CSF and plasma samples from patients in the DAT group. Although exosomes can be released by numerous organs, the expression of miR-193b in the exosomes from CSF was ~73% of the endogenous control, which indicated that it was abundant, and the exosomal miR-193b levels in the CSF were correlated. Furthermore, exosome-mediated secretion pathways exist in the blood brain barrier (29,30). It is hypothesized that decreased secretion of exosomal miR-193b may lead to the decreased level of exosomal miR-193b in the plasma.
In conclusion, these findings showed that miR-193b may function in the development of AD and exosomal miR-193b has potential as a novel, noninvasive blood-based biomarkers of patients with MCI and DAT.
Acknowledgements
This study was supported by the Natural Science Foundation of China (no. 81271924) and Research Fund for the Doctoral Program of Higher Education of China (no. 20121107110001). The authors would like to thank Dr Shuang Meng of the Chinese Center for Disease Control and Prevention, Beijing, People’s Republic of China for the vector construction and fluorescence detection.
References
|
Lewczuk P, Kamrowski-Kruck H, Peters O, et al: Soluble amyloid precursor proteins in the cerebrospinal fluid as novel potential biomarkers of Alzheimer’s disease: a multicenter study. Mol Psychiatry. 15:138–145. 2010. | |
|
Weiner MW: Dementia in 2012: Further insights into Alzheimer disease pathogenesis. Nat Rev Neurol. 2:65–66. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Singer O, Marr RA, Rockenstein E, et al: Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 8:1343–1349. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Tan L, Yu JT, Hu N and Tan L: Non-coding RNAs in Alzheimer’s disease. Mol Neurobiol. 47:382–393. 2013. | |
|
Junn E and Mouradian MM: MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 133:142–150. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Delay C, Mandemakers W and Hébert SS: MicroRNAs in Alzheimer’s disease. Neurobiol Dis. 46:285–290. 2012. | |
|
Hébert SS and De Strooper B: Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 32:199–206. 2009.PubMed/NCBI | |
|
Long JM and Lahiri DK: MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 404:889–895. 2011.PubMed/NCBI | |
|
Delay C, Calon F, Mathews P and Hébert SS: Alzheimer-specific variants in the 3′UTR of Amyloid precursor protein affect microRNA function. Mol Neurodegener. 6:702011.PubMed/NCBI | |
|
Hébert SS, Papadopoulou AS, Smith P, et al: Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 19:3959–3969. 2010.PubMed/NCBI | |
|
Blennow K, Hampel H, Weiner M and Zetterberg H: Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 6:131–144. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Rembach A, Faux NG, Watt AD, et al: AIBL research group, Changes in plasma amyloid beta in a longitudinal study of aging and Alzheimer’s disease. Alzheimers Dement. 10:53–61. 2014.PubMed/NCBI | |
|
Lewczuk P, Kamrowski-Kruck H, Peters O, et al: Soluble amyloid precursor proteins in the cerebrospinal fluid as novel potential biomarkers of Alzheimer’s disease: a multicenter study. Mol Psychiatry. 15:138–145. 2010. | |
|
Zetterberg H, Andreasson U, Hansson O, et al: Elevated cerebrospinal fluid BACE1 activity in incipient Alzheimer disease. Arch Neurol. 65:1102–1107. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Wang WX, Huang Q, Hu Y, Stromberg AJ and Nelson PT: Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2:193–205. 2011.PubMed/NCBI | |
|
Schonrock N, Ke YD, Humphreys D, et al: Neuronal microRNA deregulation in response to Alzheimer’s disease amyloid-beta. PLoS One. 5:e110702010.PubMed/NCBI | |
|
Hosseini HM, Fooladi AA, Nourani MR and Ghanezadeh F: The role of exosomes in infectious diseases. Inflamm Allergy Drug Targets. 1:29–37. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Pant S, Hilton H and Burczynski ME: The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 11:1484–1494. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Boon RA and Vickers KC: Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 33:186–192. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Wei PC, Tsai CH, Chiu PS and Lai SC: Matrix metalloproteinase-12 leads to elastin degradation in BALB/c mice with eosinophilic meningitis caused by Angiostrongylus cantonensis. Int J Parasitol. 41:1175–1183. 2011. View Article : Google Scholar | |
|
Wang WX, Rajeev BW and Stromberg AJ: The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 5:1213–1223. 2008.PubMed/NCBI | |
|
Liu CG, Xu KQ, Xu X, et al: 17Beta-oestradiol regulates the expression of Na+/K+-ATPase beta1-subunit, sarcoplasmic reticulum Ca2+-ATPase and carbonic anhydrase iv in H9C2 cells. Clin Exp Pharmacol Physiol. 34:998–1004. 2007.PubMed/NCBI | |
|
Dassow H and Aigner A: MicroRNAs (miRNAs) in colorectal cancer: from aberrant expression towards therapy. Curr Pharm Des. 19:1242–1252. 2013.PubMed/NCBI | |
|
Eulalio A, Schulte L and Vogel J: The mammalian microRNA response to bacterial infections. RNA Biol. 9:742–750. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR and Wyman SK: Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Giedraitis V, Sundelöf J, Irizarry MC, et al: The normal equilibrium between CSF and plasma amyloid beta levels is disrupted in Alzheimer’s disease. Neurosci Lett. 427:127–131. 2007.PubMed/NCBI | |
|
Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD and Remaley AT: MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 4:423–433. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
van Leuven F: Single and multiple transgenic mice as models for Alzheimer’s disease, Prog. Neurobiol. 61:305–312. 2000. | |
|
Ceruti S, Colombo L, Magni G, et al: Oxygen-glucose deprivation increases the enzymatic activity and the microvesicle-mediated release of ectonucleotidases in the cells composing the blood-brain barrier. Neurochem Int. 59:259–271. 2011. View Article : Google Scholar | |
|
Ma R, Jiang T and Kang X: Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 31:382012. View Article : Google Scholar : PubMed/NCBI |