Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin
- Authors:
- Published online on: December 24, 2015 https://doi.org/10.3892/mmr.2015.4716
- Pages: 1541-1550
-
Copyright: © Chang et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide and the principal cause of fatality among patients with cirrhosis (1). Despite recent advances in cancer treatment with respect to surgery, chemotherapy and biologics, the majority of cases of HCC remain incurable once they have become metastatic and are associated with a poor prognosis. Hepatocarcinogenesis has often been described as a multistep process involving a number of genetic alterations, eventually leading to the malignant transformation of hepatocytes (2). The advances in suitable therapies for the purpose of increasing survival rate are limited as the pathophysiological mechanisms causing HCC have not been fully elucidated. Revealing the pathological mechanism underlying HCC development and progression is indispensable for developing effective therapeutic agents.
The human transcriptome comprises not only large numbers of protein-coding messenger RNAs (mRNAs), but also a large set of non-protein coding transcripts that have structural, regulatory or unknown functions (3). Novel evidence shows that lncRNAs (long non-coding RNAs) exhibit important regulatory roles in tumorigenesis and cancer progression (4). For example, HOTAIR levels have been shown to be higher in tumor tissues than the adjacent normal tissue, and overexpression of HOTAIR correlates with the presence of metastasis. In addition, patients with high HOTAIR expression levels have been shown to have a poor prognosis (5). Forced expression of HOTAIR in cancer cells leads to altered histone H3 lysine27 methylation and gene expression, and increased metastasis (6). Maternally expressed gene 3 (MEG3) expression is markedly decreased in several primary human tumors and tumor cell lines by promoter hypermethylation (7). MEG3 is associated with meningioma progression by inducing the accumulation of p53 protein and selectively regulating p53 target gene expression. In addition, overexpression of MEG3 inhibits cancer cell proliferation and colony formation (8).
The long noncoding RNA, small nucleolar RNA host gene-growth arrest-specific 5 (GAS5), was originally identified via subtractive cDNA cloning of genes which are preferentially expressed in growth-arrested cells (9). GAS5 comprises 12 exons and encodes ten box C/D within its introns (10). The open reading frame encoded within GAS5 exons is short and thought not to encode a functional protein. This, together with the presence of a conserved 5′-terminal oligopyrimidine tract (5′-TOP), explains why GAS5-a ncRNA accumulates upon growth arrest in a range of species (11). Several studies have implicated GAS5 ncRNA in other types of cancer (12,13). GAS5 and/or its snoRNAs have been shown to be aberrantly expressed in breast cancer, head and neck squamous cell carcinoma (14–16) and glioblastoma multiforme (17), where tumor expression is reduced, and non-small-cell lung cancer, where overexpression of the snoRNAs U44, U76 and U78 occurs (18). However, the mechanisms underlying GAS5 regulation of cancer cell proliferation in HCC remain unclear.
Based on the above findings, the present study analyzed the expression of GAS5 in HCC tissues and adjacent normal tissues, and its effect on cell proliferation in HCC cells. It was demonstrated that GAS5 levels are significantly decreased in HCC tissues, and decreased expression of GAS5 indicates a poor prognosis in patients with HCC. In addition, GAS5 was shown to suppress hepatoma cell proliferation in a vimentin-dependent manner.
Materials and methods
Tissue samples, cell lines and clinical data collection
A total of 50 patients were analyzed in this study. The patients underwent HCC resection at the Department of General Surgery at the Zhongnan Hospital of Wuhan University (Wuhan, China) between February 2009 and March 2010. The study was approved by the ethics committee of Zhongnan Hospital of Wuhan University (Wuhan, China). Written informed consent was obtained from all patients. The diagnosis of HCC was histopathologically confirmed. No patients received preoperative treatment. Tumor and corresponding adjacent normal tissues were selected for each case. Normal human liver tissues were obtained >5 cm from the edge of the cancerous region. Resected tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction, or fixed in 4% paraformaldehyde for paraffin sectioning. Clinicopathological data was collected for all subjects including details of age, gender and HCC features, such as tumor size, differentiation, hepatitis B virus (HBV) infection, liver cirrhosis, serum α-fetoprotein (AFP) levels, tumor number and the presence of portal vein tumor thrombus (PVTT). Patient follow up was performed by telephone interview and questionnaire every 3 months until April 30, 2015. Overall survival (OS) was calculated from the date of the initial surgery to the date of death. Patients who succumbed to diseases not directly related to HCC were excluded from the study.
The following cell lines were used in this study: L02 normal human liver cells, and Huh7, Hep3B, HepG2, QGY-7701, MHCC97L and HCCLM9he human hepatoma cell lines. All cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), where they were characterized by mycoplasma detection, DNA fingerprinting, isozyme detection and determination of cell viability. All cells were cultured in the recommended media supplemented with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and 100 U/ml penicillin (Beyotime Institute of Biotechnology, Shanghai, China) and streptomycin (Beyotime Institute of Biotechnology) at 37°C in an incubator with a 5% CO2 atmosphere (Thermo Fisher Scientific, Inc.).
Ethics statement
Patient samples were collected and stored in the Zhongnan Hospital of Wuhan University tumor bank. Written informed consent was obtained from all of the patients. All procedures were approved by the Research Ethics Committee of Wuhan University. Patient data and samples were treated according to the ethical and legal standards adopted by the Declaration of Helsinki 2013.
RNA preparation, semiquantitative-reverse transcription and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The expression level of GAS5 was first examined by semiquantitative PCR in tissue from 6 patients with HCC. The expression of GAS5 was then analyzed by RT-qPCR in tissue from all 50 patients with HCC patients. Total RNA was extracted from tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. Quantification of total RNA was performed with a Nanodrop™ spectrophotometer (Thermo Fisher Scientific Inc.) at 260 and 280 nm. Only samples with an A260/A280 ratio between 1.8 and 2.0 were utilized for further analysis. Total RNA (5 µg) was used to synthesize first-strand cDNA using random primers and SuperScript II reverse transcriptase (Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. The 20 µl RT reactions were performed using a PrimeScript RT reagent kit (Takara Bio Inc., Wuhan, China) and incubated for 30 min at 37°C and 5 sec at 85°C, and then maintained at 4°C for 1% agarose gel electrophoresis. For qPCR, 2 µl diluted RT products were combined with 10 µl of 2X SYBR Master mix (Toyobo Co., Ltd. Osaka, Japan), 1 µl forward and reverse primers (10 µM; Table I; Beijing View Solid Biotechnology (Beijing, China), and 6 µl nuclease-free water in a final volume of 20 µl according to the manufacturer's instructions. Amplification was performed with the iQ5 quantitative PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA), under the following conditions: 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec, and 60°C for 30 sec, and 72°C for 30 sec. qPCR was conducted in triplicate, including non-template controls. Amplification of the appropriate product was confirmed by melting curve analysis following amplification. Relative expression of GAS5 and vimentin were calculated using the comparative cycle quantification (Cq) (2−ΔΔCq) method (19) with β-actin as the endogenous control to normalize the data. PCR products were run on agarose gels to determine size, and dissociation curves were subsequently utilized to examine the specificity of the RT-qPCR assay.
In situ hybridization (ISH)
ISH was used to detect GAS5 in HCC clinical specimens. A digoxigenin-UTP-labeled antisense RNA probe (Beijing View Solid Biotechnology) was derived from 234 to 478 nt of GAS5 by in vitro transcription using the DIG RNA Labeling kit (Roche Diagnostics, Indianapolis, IN, USA). The digoxigenin-UTP labeled sense RNA probe derived from 234 to 478 nt of GAS5 was employed as a negative control. ISH was performed using the ISH kit (Boster Bio-Engineering Company, Wuhan, China). The ISH-staining regions were reviewed and scored by two pathologists, the score standard for the staining intensity was as follows: 0, negative; 1, weak; 2, medium; and 3, strong. The proportion of GAS5-positive cells was scored as: 0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The total scores were calculated by combining two scores and ranged from 0 to 7. Total scores of ≥3 were defined as the high-expression group (positive group). This scoring method was simple and reproducible. Results were highly concordant between the two independent pathologists.
Immunohistochemistry
Vimentin and E-cadherin expression in primary tumor tissues and adjacent non-tumor tissues were examined using IHC. Paraffin-embedded blocks containing tumor tissues or non-tumor tissues and >70% primary tumor tissue were selected for IHC staining. Paraffin sections were cut to 4 µm, mounted on silanized slides and the preserved at 4°C. The paraffin sections were dewaxed, rehydrated and blocked with 0.3% H2O2. Tissue antigens were retrieved with a microwave oven at 95°C for 25 min and cooled to room temperature in 10 mmol sodium citrate buffer (pH 6.0). Each slide was washed with phosphate-buffered saline (PBS) and incubated overnight at 4°C with anti-vimentin (1:100, cat. no. 5741S, Cell Signaling Technologies, Inc.) or anti-E-cadherin (1:100, cat. no. 3195S, Cell Signaling Technologies, Inc.). Primary antibodies were diluted with background-reducing components (Dako, Glostrup, Denmark). Slides were then incubated with goat-anti-mouse IgG-horseradish peroxidase (HRP) (cat. no. BA1050; 1:5,000; Wuhan Boster Biological Technology, Ltd., Wuhan, China) or goat-anti-rabbit IgG-HRP (1:5,000; cat. no. BA1055; Wuhan Boster Biological Technology, Ltd.). Slides were stained for 2 min with diaminobenzidine tetrahydrochloride and then counterstained with hematoxylin. Tissue treated with antibody dilution solution was used as a negative control. All controls yielded satisfactory results. An FV1000 confocal laser-scanning microscope (Olympus, Tokyo, Japan) was used to examine the tissues. The total immunostaining score was calculated as the sum of the positive percentage and the staining intensity of the stained cells, which ranged from 0 to 6. The percent positivity was scored as follows: 0, 0–25%; 1, 26–50%; 2, 51–75%; and 3, ≥75%. The staining intensity was scored as: 0, no staining; 1, weakly stained; 2, moderately stained; and 3, strongly stained. The results of the immunohistochemical staining were scored by two experienced pathologists, who were blinded to the clinical data. A negative expression of protein was defined as a total score ≤3, and positive expression was defined as a total score ≥4.
Transfection assays
The GAS5 sequence (Gene-bank: NR_002578.2) was synthesized according to the full length GAS5 sequence (based on the GAS5 sequence) and then cloned into a pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific Inc.). The GAS5-pcDNA3.1 or empty vector was transfected into hepatoma cells. Transfection was conducted using LipJetTM in vitro DNA and siRNA Transfection kit (SignaGen Laboratories, Gaithersburg, MD, USA) according to the manufacturer's protocol. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. The empty pcDNA3.1 vector was used as the control. All transfection experiments were performed at least three times.
Fluorescent immunocytochemistry
Hepatoma cells were fixed with 4% paraformaldehyde [China Sinopharm International (Shanghai), Co., Ltd.] for 20 min and then permeabilized with 0.5% Triton X-100 [China Sinopharm International (Shanghai), Co., Ltd.] for 10 min. The fixed cells were blocked with 3% FBS for 30 min. For specific detection of vimentin protein (Cell Signaling Technologies, Inc., Boston, MA, USA), the cells were incubated with polyclonal rabbit anti-rat vimentin antibody (1:100, CST, USA) overnight at 4°C. The samples were incubated with Cy3-labeled goat anti-rabbit IgG (1:100, eBioscience, Inc., San Diego, CA, USA) in PBS for 1 h at 37°C. Then, the cells were washed twice with PBS and incubated with 4′, 6-diamidino-2-phenylindole (DAPI; BD Biosciences, La Jolla, CA, USA) for 5 min. The images were analyzed using fluorescence microscopy (Nikon Eclipse 80i, Tokyo, Japan). Identical illumination and camera settings were used within each dataset.
Cell proliferation assay
Cell proliferation assays were performed using Cell Counting kit-8 (Dojindo Molecular Technologies Inc., Kumamoto, Japan), according to the manufacturer's protocol. Human hepatoma cells were plated in 24-well plates in triplicate at a density of 2-5×104 cells per well and cultured in growth medium. Cells were then treated with pcDNA3.1, empty vector or vimentin-small interfering (si)RNA (Beijing View Solid Biotechnology) and the numbers of cells per well were measured by the (450 nm) at the indicated time points. The plasmid GAS5-pcDNA3.1 (Beijing View Solid Biotechnology) was constructed by introducing a KpnIXhoI fragment containing the GAS5 cDNA into the same sites in pcDNA3.1.
Bromodeoxyuridine (Brdu) assay
Hepatoma cells were fixed in 4% paraformaldehyde for 20 min, cells were then washed three times in PBS, dyed with BrdU (BD Biosciences) for 30 min, washed again in PBS three times, and treated with DAPI for 5 min. The images were analyzed using fluorescence microscopy (Nikon Eclipse 80i). Identical illumination and camera settings were used within each dataset.
Flow cytometric analysis
Hepatoma cells (2-5×105) treated with GAS5-pcDNA3.1 or negative control (NC) were plated in 6-well plates. After 48 h incubation the cells were harvested by trypsinization. The cultures were double stained with incubated with Annexin V and propidium iodide [China Sinopharm International (Shanghai), Co., Ltd.] for 30 min in the dark. Cultures were collected and analyzed for cell apoptosis using a flow cytometer (FACScan; BD Biosciences Franklin Lakes, NJ, USA) equipped with CellQuest 3.3 software. Cells were categorized as early apoptotic cells, late apoptotic cells, dead cells, or viable cells. The ratio of early apoptotic cells was compared with that in the controls from each experiment.
Cell invasion assays
After transfection for 24 h, cells in serum-free media were seeded into the upper chamber of a Transwell apparatus (BD Biosciences). for invasion assays with Matrigel (Sigma-Aldrich, St. Louis, MO, USA). The lower chambers were filled with media containing 10% FBS. After 24 h of incubation at 37°C in a 5% CO2 atmosphere, the cells which had invaded through the membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma-Aldrich). The cells on the lower surface were photographed (Olympus; IX73) and three random fields were counted using a confocal laser-scanning microscope (Olympus FV1000). Three independent experiments were performed.
Western blot analysis
Western blot analysis to assess vimentin and E-cadherin protein expression was performed as previously described (20). Glyceraldehyde 3-phosphate dehydrogenase served as a control. Mouse monoclonal anti-GAPDH (cat. no. KM9002; 1:5,000; Sungene, Tianjin, China), rabbit monoclonal anti-vimentin (cat. no. 5741S; 1:1,000; Cell Signaling Technology, Inc.) and rabbit monoclonal E-cadherin (cat. no. 3195S; 1:1,000; Cell Signaling Technology, Inc.) primary antibodies were used. Goat anti-mouse IgG-HRP (cat. no. BA1050; 1:5,000; Boster Bio-Engineering Company) and goat anti-rabbit IgG-HRP (cat. no. BA1055; 1:5,000, Boster Bio-Engineering Company).
Statistical analysis
All statistical analyses were performed using SPSS software (SPSS, Inc., Chicago, IL, USA). The differences between groups were tested using Student's t-test. Survival curves were calculated using Kaplan-Meier and log-rank tests. The effects of variables on survival were determined by univariate and multivariate Cox proportional hazards modeling. The correlation between GAS5 and vimentin was analyzed by Spearman's rank correlation. Two-sided P-values were calculated. P<0.05 was considered to indicate a statistically significant difference.
Results
GAS5 is significantly downregulated in HCC tissues and hepatoma cells
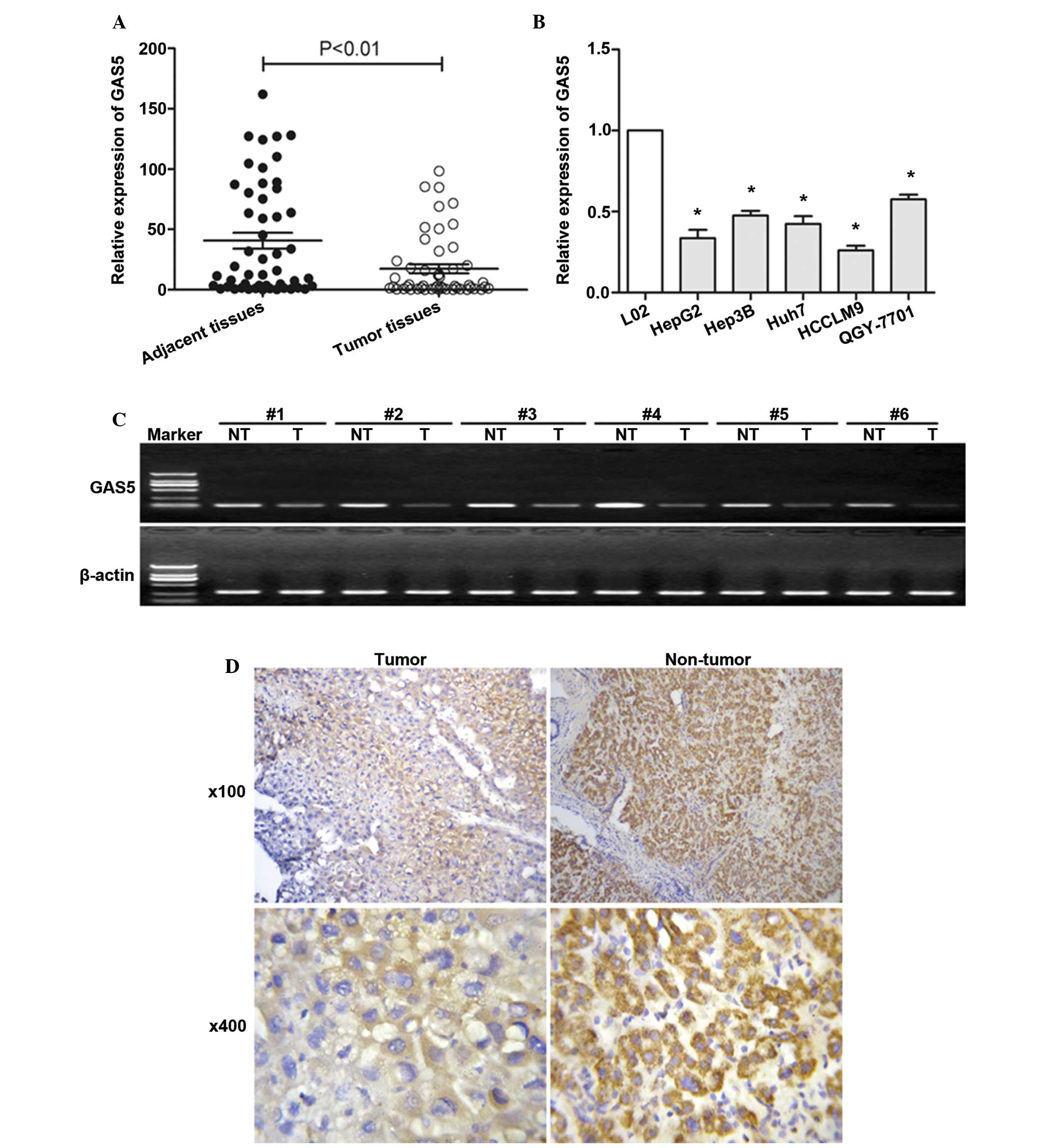
To investigate whether GAS5 regulates hepatocarcinogenesis, the expression level of GAS5 was examined in HCC and adjacent normal tissues (Fig. 1). The expression level of GAS5 was first examined by semiquantitative RT-PCR in tissue from 6 patients with HCC. GAS5 was downregulated in HCC tissues compared with adjacent non-tumor tissues (Fig. 1C). The expression of GAS5 was then investigated by RT-qPCR in tissue from all 50 patients with HCC. Fig. 1A showed significantly lower GAS5 expression in HCC tissues. Fig. 1A and C show that GAS5 is significantly downregulated in HCC tissues compared with adjacent normal tissues. The mean value of GAS5 expression in normal tissues was 40.74 but the mean value of GAS5 expression in HCC tissues is 17.35. GAS5 expression levels were also assessed in hepatoma cells, as shown in Fig. 1B, GAS5 was observed to be downregulated in hepatoma cells compared with normal liver cells. An ISH assay (Fig. 1D) further confirmed that GAS5 was significantly downregulated in HCC tissues compared with adjacent normal tissues.
GAS5 expression and clinicopathological factors in HCC
As shown in Table II, in order to assess the correlation between GAS5 expression and clinicopathological data, tumor tissues were divided into the low-expression group (mean expression value 1.24, n=25) and the high-expression group (mean expression value 33.45, n=25), based on the mean expression level of all tumor tissues (mean expression value, 2.98). Clinicopathological factors were analyzed in the high and low GAS5 expression groups. The patients with low GAS5 (n=25) exhibited a poorer histologic grade (P<0.010) and increased presence of PVTT (P=0.001) than the high GAS5 expression group (n=25). However, there was no significant correlation between GAS5 expression and other clinicopathologic features, such as age, gender, HBV infection, liver cirrhosis, serum AFP and tumor number (P>0.05).
Low expression of GAS5 indicates a poor prognosis in HCC
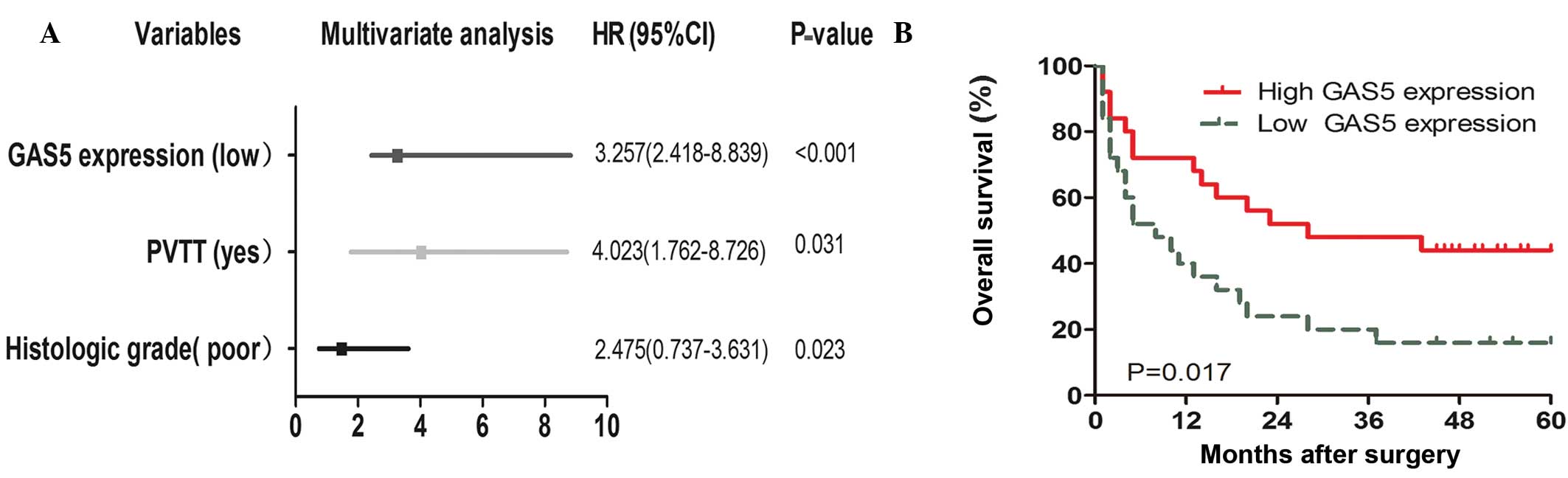
To determine the factors responsible for patient survival, univariate and multivariate analysis were performed. GAS5 expression levels were obtained from the RT-qPCR data of the cohort of 50 patients mentioned above. Univariate analysis of overall survival revealed that GAS5 expression (P=0.000), PVTT (P=0.008), and differentiation (P=0.003) were prognostic indicators (Table III). Multivariate analysis showed that GAS5 expression was an independent prognostic indicator for overall survival (P<0.001; Fig. 2A). No significant associations were found with age, gender, HBV infection, liver cirrhosis, serum AFP and tumor number (Table III). Furthermore, high expression of GAS5 was found to be associated with lower rates of OS (P=0.017; log-rank test; Fig. 2B). However, no significant associations were found between OS and age, gender, HBV infection, liver cirrhosis, serum AFP and tumor number.
Table IIIUnivariate and multivariate analysis using the Cox proportional hazard regression model for overall survival. |
GAS5 regulates cell apoptosis and cell proliferation in vitro
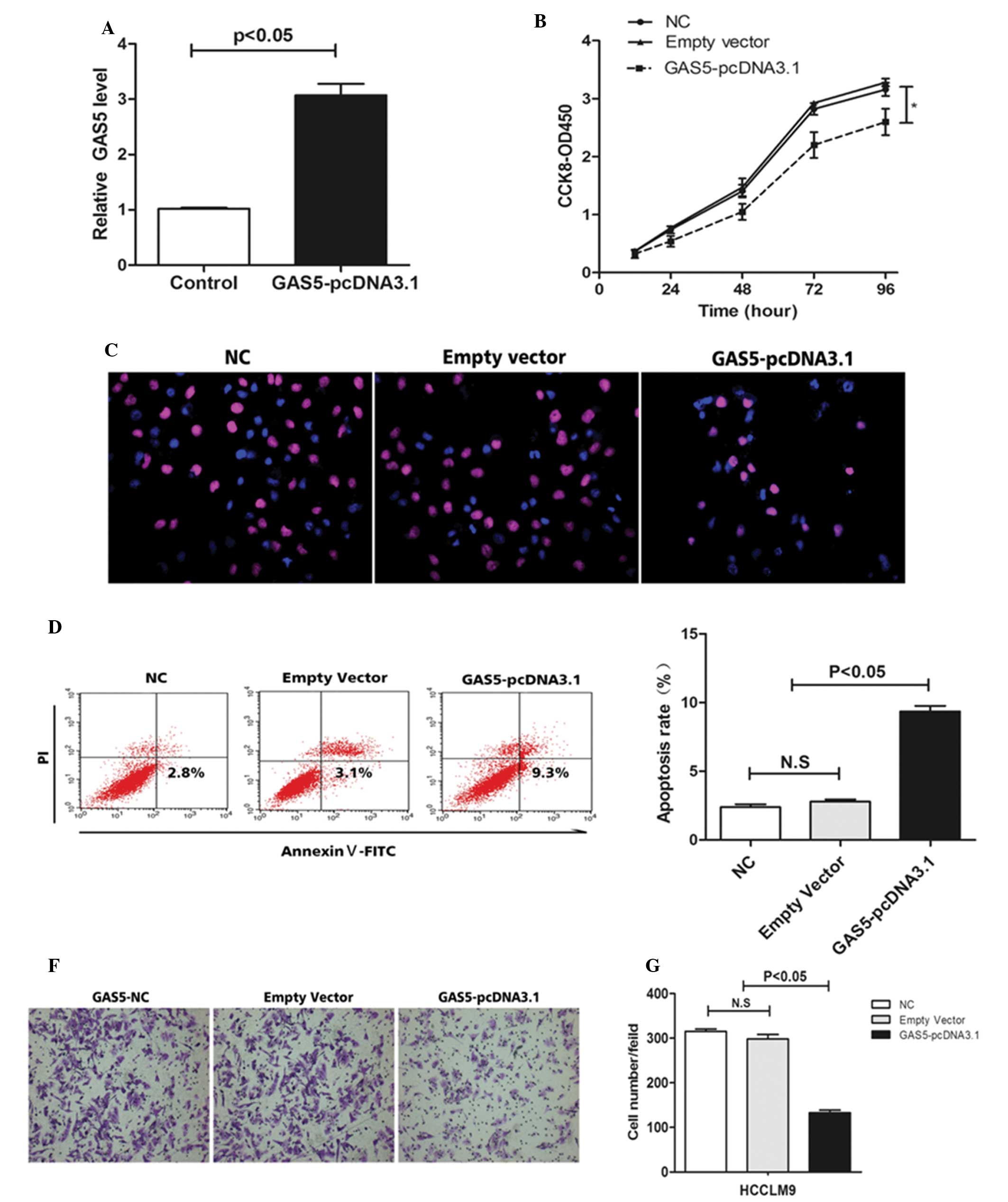
To investigate the role of GAS5 in the regulation of cell proliferation, cell apoptosis and the ability of cell invasion, the hepatoma cells were treated with GAS5-pcDNA3.1. RT-qPCR was used to observe the expression of GAS5 (Fig. 3A). Overexpression of GAS5 inhibited HCCLM9 proliferation (Fig. 3B and C) and promoted HCCLM9 apoptosis (Fig. 3D and E). In addition, overexpression of GAS5 suppressed the invasion ability of HCCLM9 (Fig. 3F and G). The above data demonstrated that GAS5 is key in hepatocarcinogenesis.
Expression of vimentin and E-cadherin in HCC
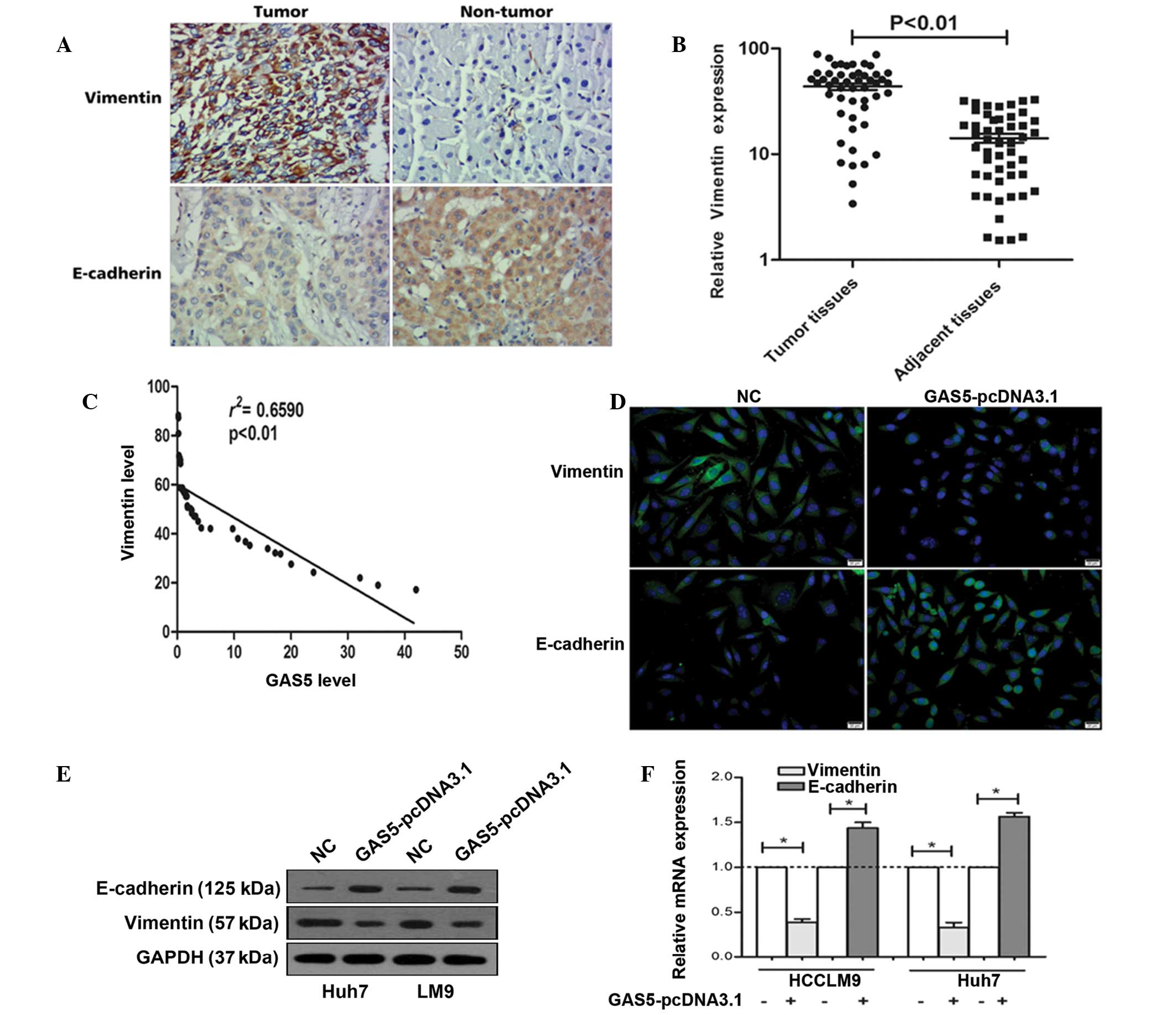
Epithelial-mesenchymal transition (EMT) has been shown to be of critical importance in the early events of tumor cell metastatic dissemination as cells become more motile and acquire invasive potential (21). It was previously found that GAS5 suppressed the invasion ability of tumor cells. Thus, the present study determined the expression of vimentin and E-cadherin (marker protein of EMT) (22). As shown in Fig. 4A, immunohistochemistry indicated that vimentin was significantly upregulated in tumor tissues, while E-cadherin was downregulated in tumor tissues. In addition, RT-qPCR confirmed the above phenomenon (Fig. 4B).
GAS5 negatively regulates vimentin expression in vitro
To investigate the correlation between GAS5 and vimentin, Spearman's rank correlation analysis was used. As shown in Fig. 4C a significant negative correlation is observed between the GAS5 levels and the vimentin levels in HCC tissues (r2=0.6590, P<0.01). In addition, fluorescent immunocytochemistry indicated that overexpression of GAS5 decreased vimentin expression and increased E-cadherin expression (Fig. 4D). Western blot analysis demonstrated that the vimentin protein level is downregulated and E-cadherin is upregulated in Huh7 and HCCLM9 after GAS5 overexpression (Fig. 4E). Figure 4F shows that overexpression of GAS5 significantly decreased the vimentin mRNA levels and increased E-cadherin mRNA in Huh7 and HCCLM9 cells. These data indicated that GAS5 regulates hepatoma cell proliferation and invasion by the regulation of vimentin.
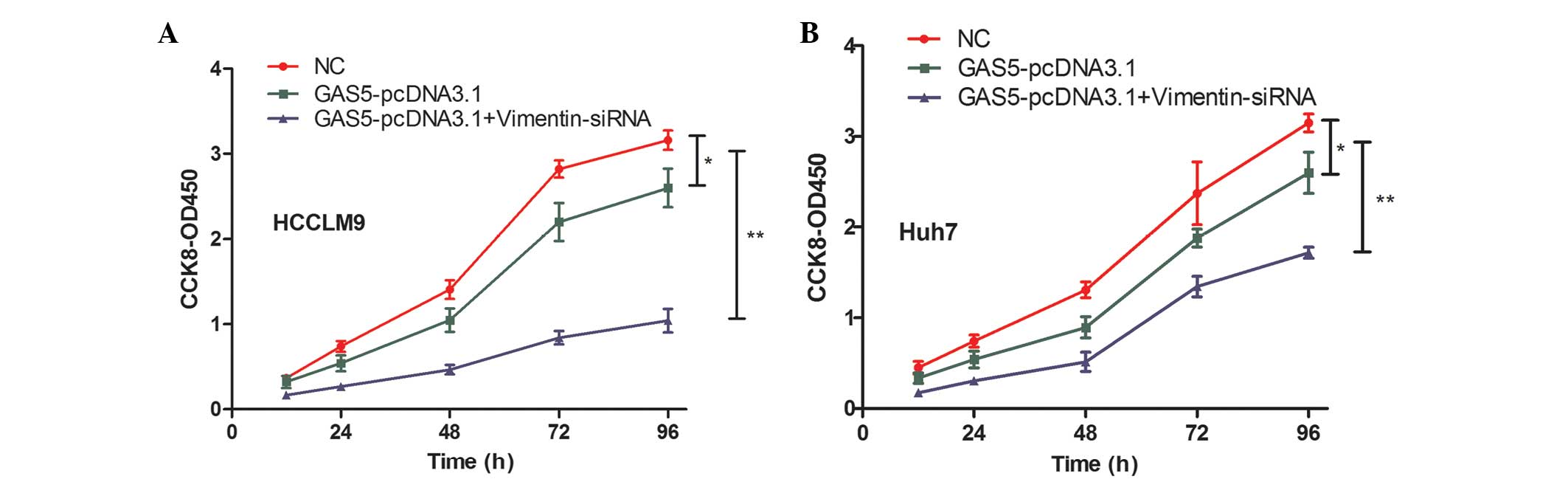
GAS5 inhibits cell proliferation by regulating vimentin
GAS5 overexpression increased hepatoma cell proliferation and a significant negative correlation was observed between GAS5 and vimentin. Thus, it was hypothesized that the role of GAS5 in regulating hepatoma cell proliferation is mediated by vimentin. Figure 5A shows that vimentin knockdown partially strengthened the ability of GAS5-pcDNA3.1-inhibited cell proliferation in HCCLM9 cells. Vimentin inhibition also strengthened the ability of GAS5-pcDNA3.1-inhibited cell proliferation in Huh7 cells (Fig. 5B). These data confirmed that GAS5 suppressed hepatoma cell progression, at least in part, by regulating vimentin expression.
Discussion
In recent years, genome-wide surveys have revealed that 98% of the total human genome can be transcribed, including certain short or lncRNAs with numbered or no protein-coding capacity (23–25). Up to now, studies have identified a huge number of lncRNAs involved in the development and progress of human diseases, including cancer (21,26). LncRNAs can function as regulators of tumor suppressor or oncogene expression and may be one of the 'cutt' that lead to oncogenesis (27).
To date, there is increasing evidence that suggests that certain lncRNAs can be identified as biomarkers for the prognosis of tumor therapeutic targets in human cancer (28). GAS5 is an lncRNA involved in the regulation of cell cycles. GAS5 is key in normal growth arrest in T-cell and non-transformed lymphocytes. GAS5 inhibition suppresses cell apoptosis and maintains a more rapid cell cycle, whereas GAS5 overexpression results in an increase in cell apoptosis and a reduction in the rate of progression through the cell cycle, revealing that GAS5 is required for normal growth arrest (14). Recently, studies have identified a novel role of GAS5 in the regulation of tumorigenesis (29). GAS5 was downregulated in a number of tumor types, such as breast cancer, head and renal cell carcinoma, prostate cancer, and glioblastoma (30–32). These observations indicate that GAS5 may function as a tumor suppressor in human tumor progression. However, the molecular mechanisms underlying GAS5 regulation of cancer cell proliferation in HCC remain unclear.
EMT has been shown to be of critical importance in the early events of tumor cell metastatic dissemination during which cells become more motile and exhibit invasive potential (33). EMT is not just a mechanism that forms fibroblast-like cells; it is a process that results in cancer cell migration, invasion, and metastasis (34,35). Various biomarkers have been screened to indicate the EMT process, including the loss of E-cadherin, zona occludens-1 and cytokeratin, and the upregulation of matrix metalloproteinase, fibronectin, α-smooth muscle actin, vimentin, snail and slug (35). Vimentin is a 57 kDa, type III intermediate filament that is found in mesenchymal cells of various types of tissues during their developmental stages. Its function is to maintain cell and tissue integrity. Vimentin is associated with tumor invasion and a poor prognosis in a number of types of cancers, including breast cancer, prostate cancer, melanoma and lung cancer, and serves as a potential target for cancer therapy (36,37).
The current study verified that the expression level of GAS5 is significantly downregulated in HCC tissues compared with adjacent normal controls. Decreased expression of GAS5 was associated closely with the tumor differentiation (P<0.010) and PVTT (P=0.001). The downregulated expression of GAS5 was associated with poor prognosis of HCC. GAS5 overexpression decreases hepatoma cell proliferation and invasion. In addition, GAS5 overexpression promotes the apoptosis of hepatoma cells. It was further demonstrated that GAS5 is involved in the EMT of HCC cells. Overexpression of GAS5 downgegulated the vimentin level and the upregulated E-cadherin level in hepatoma cells. A significant negative correlation was observed between GAS5 and the vimentin level in vivo. Notably, knockdown of vimentin partially increased GAS5-pcDNA3.1-induced inhibition of cell proliferation.
These data suggest an important role of GAS5 in the molecular etiology of HCC and implicate the potential application of GAS5 in HCC therapy.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant nos. 81472268/H1617 and 81272692/H1617).
References
|
Venook AP, Papandreou C, Furuse J and de Guevara LL: The incidence and epidemiology of hepatocellular carcinoma: A global and regional perspective. Oncologist. 15(Suppl 4): S5–S13. 2010. View Article : Google Scholar | |
|
El-Serag HB and Rudolph KL: Hepatocel lu la r carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al: Long noncoding RNAs with enhancer-like function in human cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gibb EA, Brown CJ and Lam WL: The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 10(38)2011. View Article : Google Scholar : PubMed/NCBI | |
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al: Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou Y, Zhang X and Klibanski A: MEG3 noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Schneider C, King RM and Philipson L: Genes specifically expressed at growth arrest of mammalian cells. Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI | |
|
Smith CM and Steitz JA: Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 18:6897–6909. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Williams GT and Farzaneh F: Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 12:84–88. 2012.PubMed/NCBI | |
|
Yu X and Li Z: Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 10:1953–1958. 2015.PubMed/NCBI | |
|
Glover AR, Zhao JT, Ip JC, Lee JC, Robinson BG, Gill AJ, Soon PS and Sidhu SB: Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr Relat Cancer. 22:99–109. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 28:195–208. 2009. View Article : Google Scholar | |
|
Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z and Jiang F: Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 9(198)2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al: The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 104:1168–1177. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al: Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z and Jiang F: Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 9(198)2010. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan Q, Loya K, Rani B, Möbus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, Cantz T and Sharma AD: MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 57:299–310. 2013. View Article : Google Scholar | |
|
Kapinas K, Kessler C, Ricks T, Gronowicz G and Delany AM: miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 285:25221–25231. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Takahashi K, Yan I, Haga H and Patel T: Long noncoding RNA in liver diseases. Hepatology. 60:744–753. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q and Zhang J: Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. Sept 24–2015.Epub ahead of print. | |
|
ENCODE Project Consortium; Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al: Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Amaral PP, Dinger ME, Mercer TR and Mattick JS: The eukaryotic genome as an RNA machine. Science. 319:1787–1789. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Louro R, Smirnova AS and Verjovski-Almeida S: Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar | |
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI | |
|
Taft RJ, Pang KC, Mercer TR, Dinger M and Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol. 220:126–139. 2010. View Article : Google Scholar | |
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Tani H, Torimura M and Akimitsu N: The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 8:e556842013. View Article : Google Scholar : PubMed/NCBI | |
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Cao S, Liu W, Li F, Zhao W and Qin C: Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 7:6776–6783. 2014.PubMed/NCBI | |
|
Pickard MR, Mourtada-Maarabouni M and Williams GT: Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Thiery JP, Acloque H, Huang RY and Nieto MA: Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Kalluri R and Weinberg RA: The basics of epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Zeisberg M and Neilson EG: Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Satelli A and Li S: Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Lehtinen L, Ketola K, Makela R, Mäkelä R, Mpindi JP, Viitala M, Kallioniemi O and Iljin K: High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 4:48–63. 2013. View Article : Google Scholar : PubMed/NCBI |