Regulation of M1‑type and M2‑type macrophage polarization in RAW264.7 cells by Galectin‑9
- Authors:
- Published online on: October 4, 2017 https://doi.org/10.3892/mmr.2017.7719
- Pages: 9111-9119
Abstract
Introduction
Macrophages are involved in the occurrence, development and digestion of inflammation and other stages. At different stages of the inflammatory response, macrophages exhibit different phenotypes (1). Macrophages have at least two different polarizations, namely M1-type polarization and M2-type polarization (1,2). The former is characterized by elevation of pro-inflammatory cytokines, antimicrobial and tumoricidal activity, whereas the latter is linked to immunosuppression and tissue repair (1,2).
Activation of M1-type macrophages is typically driven by Thl-type cytokine interferon-Y, bacterial-related components such as Toll-like receptor (TLR) analogs (1,3). Type M1 is also characterized by efficient generation of reactive oxygen intermediates (ROI), inflammatory cytokines such as IL-1β, TNF and IL-6, and cytotoxicity (phagocytosis of microorganisms and necrotic cells) (4). In mice, M1 macrophage-associated markers include IL-12, MHC class II molecules and nitric oxide synthase 2 (NOS2) (5,6).
More and more studies had shown that macrophage polarization imbalance is the key pathological factors of a variety of immune-related diseases such as autoimmune diseases, tumors and other diseases (7). The study of target molecules for macrophage polarization regulation has become a new research hotspot. Although it is known that signal molecules such as signal transducer and activator of transcription (Stat)1, Stat3, Stat6, SOCS1, IRF4 and IRFS, as well as a variety of miRNAs, are involved in the macrophage polarization regulation (8,9), but different receptor molecules how to activate the polarization regulation signal above-mentioned is not very clear. It is of great theoretical and practical significance to find a new type of macrophage regulatory molecule and to clarify its regulatory mechanism for the targeted immune intervention of macrophages.
Galectin is a family of lectin that specifically binds to the glycoprotein-β-galactose residue and is present in a wide variety of organisms and is widely distributed in the nucleus, cytoplasm and extracellular matrix (10). Galectin has a variety of biological functions, such as cell adhesion, cell growth regulation, apoptosis, inflammatory response, immune regulation (10,11). Recent report had showed that macrophages with upregulated Galectin-3 participate in liver cirrhosis through production of both M1- and M2-related factors (12). Another report revealed that the inhibition of Gal-3 binding to integrin promoted macrophages phenotype towards the M1 phenotype (13). Gal-9 is also a member of the Galectin family and has a typical conserved region of the Galectin family (14). It is widely distributed in the liver, lung, tonsil, islet cells and various immune cells (14). Gal-9 plays an important role in the regulation of various diseases, including T cell-mediated diseases (such as autoimmune diseases and asthma), tumors (such as melanoma, cervical cancer, hepatic carcinoma) (13,14). However, these biological effects of Gal-9 are mostly activated by their binding to T cell immunoglobulin mucin (Tim)-3 (14,15). Tim-3 is an important member of the Tim gene family, mainly expressed on the surface of activated Th1 cells, and plays an important immunomodulatory role when combined with its ligand Gal-9 (14–16). Recently, the study found that Tim-3 is also expressed in natural immune cells (17). In addition, investigations showed that the expression of Tim-3 in sepsis is related to the overactivation of macrophages in vivo (18), suggesting that Tim-3 may be a new regulatory target for macrophages via changing the levels of Galectin-9 (Gal-9). However, the effect of Gal-9 on macrophage polarization is poorly understood to date.
Since the polarization state of macrophages has a crucial effect on its own function, and the mechanism of regulation of polarization is not yet clear. Therefore, the aim of this study was to investigate whether Gal-9 is involved in the regulation of macrophage polarization.
Materials and methods
Cell culture
The mouse macrophage line RAW264.7 is a product of ATCC. Cells were cultured in DMEM medium supplemented with 5% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin and placed in a cell culture chamber at 37°C in a humidified atmosphere with 5% CO2. After the cells were grown to 80% confluence, the culture medium was removed and washed with PBS and digested with 0.25% trypsin. Logarithmic growth phase cells were used for experiments as follows.
Cell transfection
Gal-9 siRNA (Santa Cruz Biotechnology, Dallas, TX, USA) and recombinant plasmid containing Gal-9 (Promocell, Heidelberg, Germany) were transfected into RAW264.7 cells with Invitrogen Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Briefly, cells were seeded in 6-well plates and grown to 60 to 70% confluence. Cells were then suspended with transfection reagent and Gal-9 specific siRNA (or scramble siRNA) or Gal-9 recombinant plasmid (or mock plasmid) were added with a concentration of 10 nM into a 1.5 ml centrifuge tube and incubated for 10 min. Lipofectamine 2000 (10 µl) was added to another 1.5 ml centrifuge tube and incubated at room temperature for 10 min. The two tube solutions were then mixed and incubated at room temperature for 20 min. The mixed solution was then added to 6 wells and placed into the incubator at 37°C, 5% C02 for 5 h. After transfection for 5 h, the transfected cells were incubated for another 24 h in complete medium. The transfected cells were divided into several groups as follows: Control (without treatment), Scramble, pGal-9, mock, (negative siRNA), Gal-9-siRNA and then the expression levels of Gal-9 mRNA and protein were performed. After transfected for 48 h, the cells were harvested to conduct analysis performed as follows.
Detection of cell proliferation activity by CCK-8 assay
Cell proliferation activities from all groups were determined by CCK-8 assay after cells transfected for 0, 6, 12, 24, 48 h. Briefly, cells were inoculated in 96-well plates at 1×105 cells per well. CCK-8 (20 µl; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was then added and the cells were incubated for another 4 h. The absorption of cell solutions was read at 450 nm using an ELISA reader (BioTek Instruments, Inc., Winooski, VT, USA) following the manufacturer's instructions.
Detection of cell polarization by flow cytometry
Cells collected from all groups were fixed with 4% paraformaldehyde at room temperature for 30 min and washed twice with PBS, and then punched with 0.3% Triton X-100 for 5 min. After washed twice with PBS, cells were blocked with 1% BSA 200 ml for 1 h. Cells solutions were then centrifuged and the supernatants were discarded. Then, 0.125 µg of FITC-labeled CD206 and 0.25 µg of PE-labeled NOS2 (both from BioLegend, Inc., San Diego, CA, USA) were added, respectively, and incubated at 40°C for 30 min. Followed by washing twice with PBS, and the cell polarity was measured by flow cytometry.
Enzyme-linked immunosorbent assay (ELISA) assay
Cell supernatant was isolated from all groups after centrifugation at 14,000 rpm at 4°C for 20 min. Cell (20 µl) supernatant from all groups were transferred to a 96-well plate and NOS2, TNF-α, CD206, TGF-β levels were measured using specific ELISA kits (Biosource International Inc., Camarillo, CA, USA). Briefly, the ELISA assays were conducted using 96-well microplate coated with anti-NOS2, TNF-α, CD206, TGF-β monoclonal antibody. Samples and NOS2, TNF-α, CD206, TGF-β standard controls were added to the plate, and then incubated at room temperature with shaking for 2 h and 6 washes. The concentration of Gal-9 in supernatant was also measured by a commercial ELISA kit (Cygnus, Southport, NC, USA) according to manufacturer's instructions. Chemiluminescent signal was subsequently measured at 450 nm.
Dual-luciferase reporter gene assay
RAW264.7 cells were seeded on 24-well plates at a density of 8X104 cells/well. To detect the transcription activity of NF-κB, RAW264.7 cells were co-transfected with firefly luciferase reporter gene, rennet luciferase reporter gene, and Gal-9 plasmid with 1 ml Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). After incubation for 48 h, the individual samples were collected and the cells were lysed with the PLB lysate in the kit, and the protein was extracted and used to detect luciferase activity. The substrate was added and the activity of luciferase was determined. The change in NF-κB activity in RAW264.7 cells was reflected by the ratio of firefly luciferase activity/bloody luciferase activity.
Western blot assay
The cell lysates from all groups were extracted using lysis Triton X-100 buffer containing 50 mM Tris (pH 7.6), 1% Triton X-100, 150 mM NaCl, 0.1% sodium dodecylsulphate (SDS), 1% deoxycholate, 0.2% aprotinin and1 mM PMSF (Sigma-Aldrich, St. Louis, MO, USA). Protein content was detected using the BCA protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). The equal protein samples were mixed into the wells of 7.5% polyacrylamidegels. The samples were then resolved by 10% SDS-PAGE gel and transferred on PVDF membrane (Roche Diagnostics, Penzberg, Germany). Membranes were then blocked with 10% non-fat dried milk in PBS. The blots were then incubated with anti-p-Stat1, anti-Stat1, anti-p-Stat3 anti-Stat3, anti-Gal-9 antibody at 4°C overnight. β-actin was used as an internal control. After washed with PBS, the blots were incubated with appropriate HRP-conjugated secondary antibodies at room temperature for 1 h. The blots were visualized by using chemiluminescence and developed onto the film (Kodak, Rochester, NY, USA) according to the manufacturer's instructions.
RT-PCR assay
Total cellular RNA was isolated from all groups using the RNeasy mini kit (Qiagen, Inc., Valencia, CA, USA) according to manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA using the Omniscript™ Reverse Transcriptase kit (Qiagen, Inc.). Real-time PCR was performed using a SYBR-Green PCR Master mix (Applied Biosystems, Foster. City, CA, USA). Analysis was performed on an ABI PRISM 7700 Sequence Detection system (Applied Biosystem) according to the manufacturer's protocol. The specificity of PCR products obtained was verified by melting curve analysis and agarose gel electrophoresis. The expression of Gal-9, NOS2, TNF-α, CD206 and TGF-β1 were normalized against to β-actin and were calculated using delta-delta cycle threshold (CT) method. Primers were designed by using ‘Primer3’ on website. Briefly, we acquired the primers on website ‘Primer3 web version 4.0.0’ according to the gene sequences obtained from ‘NCBI’. Primer sequences for Gal-9, NOS2, TNF-α, CD206, TGF-β are listed as follows: Gal-9, 5′GGGCAGGAAGAGCGAAGTCT3′ (forward) and 5′GCTGGATATCACCCGCCACT3′ (reverse); TNF-α, 5′CCAGGCAGGTTCTGTCCCTT3′ (forward) and 5′ATAGGCACCGCCTGGAGTTC3′ (reverse); IL-10, 5′CCAGTACAGCCGGGAA GACA3′ (forward) and 5′GAAGGCAGTCCGCAGCTCTA3′ (reverse); IL-6, 5′CTGGAGCCCACCAAGAACGA3′ (forward) and 5′GCCTCCGACTTGTGAAGTGGT3′ (reverse); TGF-β1, 5′GCCACTGCCCATCGTCTACT3′ (forward) and 5′CACTTGCAGGAGCGCACAAT3′ (reverse); β-actin, 5′GCCGGGACCTGACAGACTAC3′ (forward) and 5′TGGCCATCTCCTGCTCGAAG3′ (reverse).
Statistical analysis
The data from three independent experiments was expressed as means ± standard deviation. Statistical comparisons between different groups were calculated by SPSS software (SPSS version 20; IBM SPSS, Armonk, NY, USA). P<0.05 was considered statistically significant.
Results
Recombinant Gal-9 and siRNA of Gal-9 are effectively transfected into RAW264.7 cells
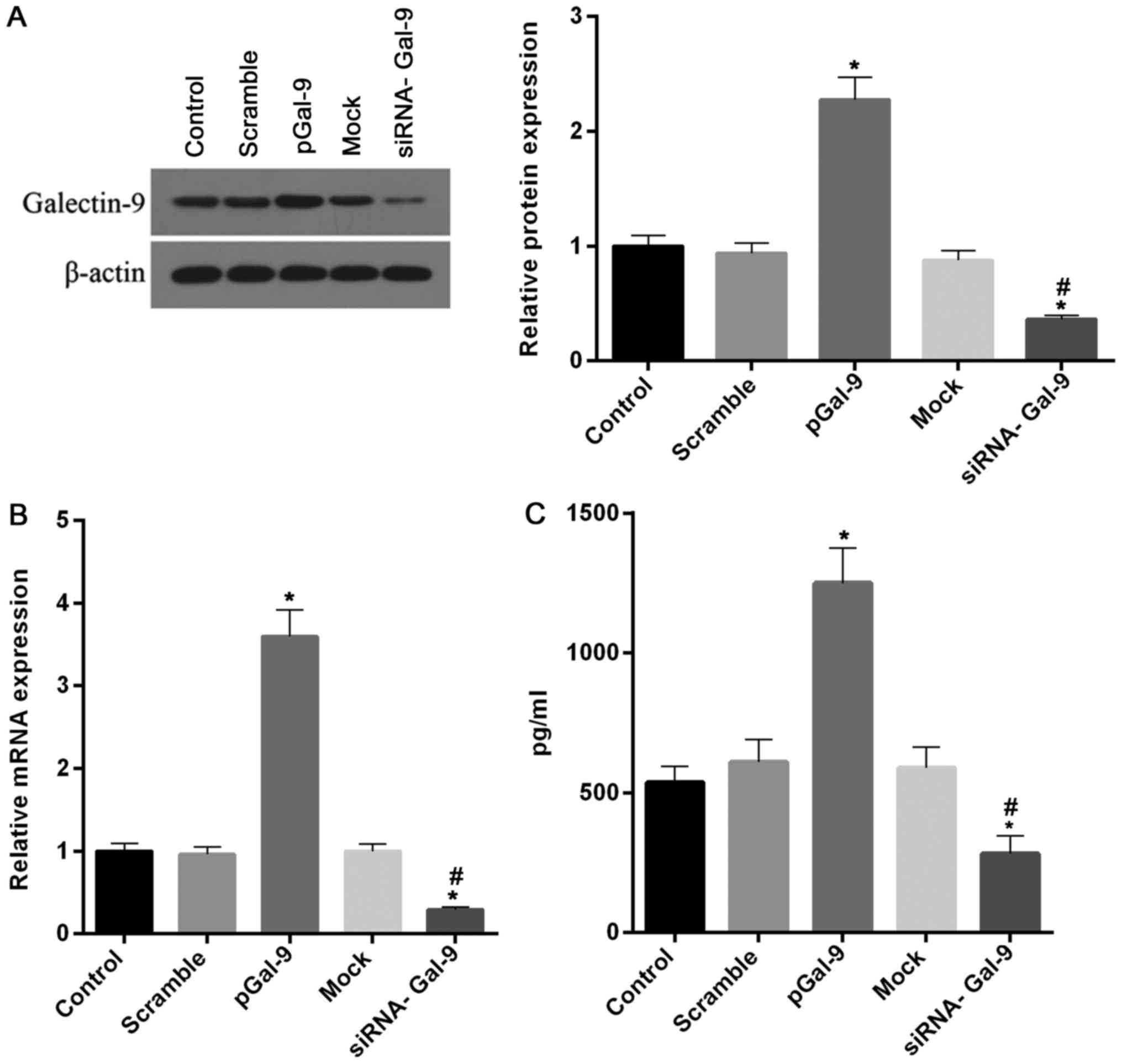
In order to explore different expression levels of Gal-9 on the regulation of macrophage polarization, we constructed cell models transfected with recombinant Gal-9 genes or siRNA targeting Gal-9. After cells transfected for 48 h, the expression of Gal-9 was tested to confirm the transfection effectiveness. Our results revealed that highly effective transfection of recombinant Gal-9 and siRNA of Gal-9 were obtained in RAW264.7 cells. As showed in Fig. 1, the levels of Gal-9 protein and mRNA were markedly increased and decreased in cells treated with transfection of recombinant Gal-9 (pGal-9) and siRNA of Gal-9 (siRNA-Gal-9), respectively, compared to cells from control, negative control of recombinant Gal-9 (Scramble) or siRNA of Gal-9 (Mock). Moreover, we tested the concentration of Gal-9 in supernatant. The profile of Gal-9 concentration in the supernatant is consistent with the intracellular level of Gal-9 protein (Fig. 1C).
Cell viability of cells is reduced in cells transfected with siRNA-Gal-9
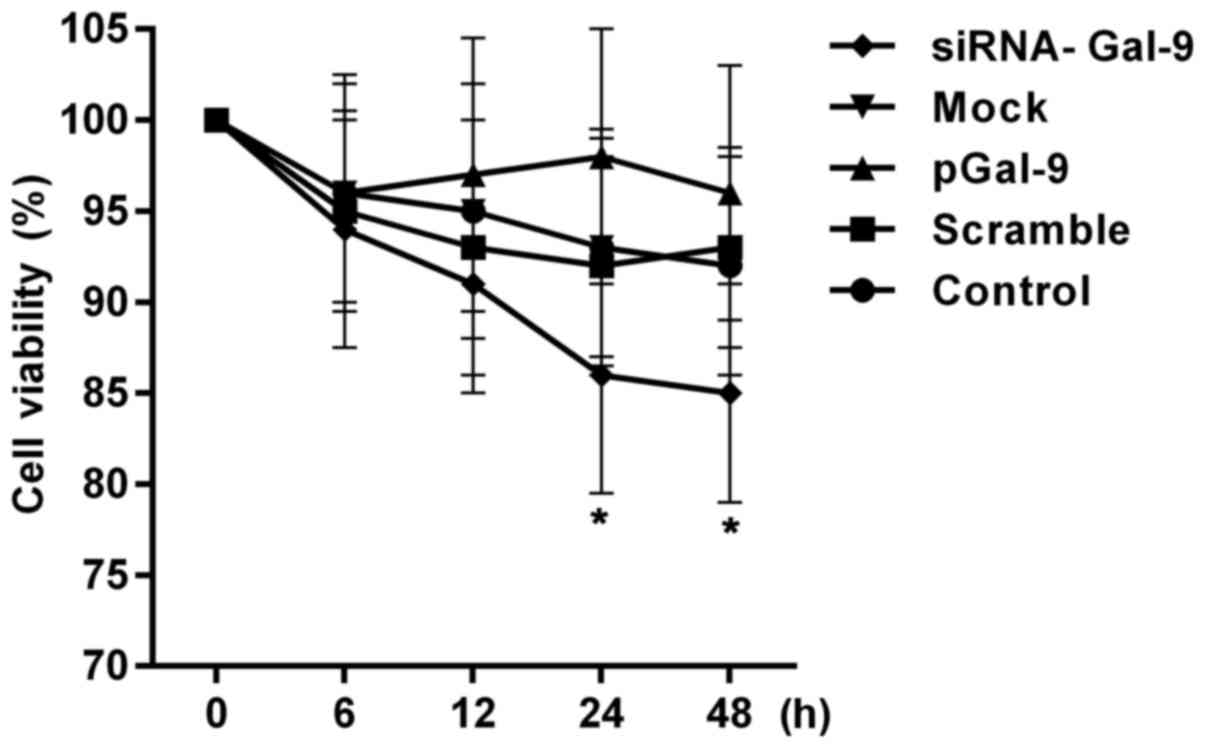
To assess the effects of transfection treatment on the cell proliferation activity, we detected the cell viabilities. Fig. 2 showed a CCK-8 assay of relative cell viability of cells at 0, 6, 12, 24, 48 h after cell transfection. The cell viabilities were altered in cells from pGal-9 and siRNA-Gal-9 group compared to Control, Scramble and Mock group. The cell viability of cells from siRNA-Gal-9 was apparent lower than that of the others, especially 24 h after the transfection. Although a slight higher of cell viability was observed in cells from pGal-9 group compared to Control, Scramble and Mock group, there was no significant difference between them. However, the cell viabilities were nearly equal among Control, Scramble and Mock group.
Expression levels of NOS2 and CD206 in cells are altered in the opposite way after regulation of Gal-9 level
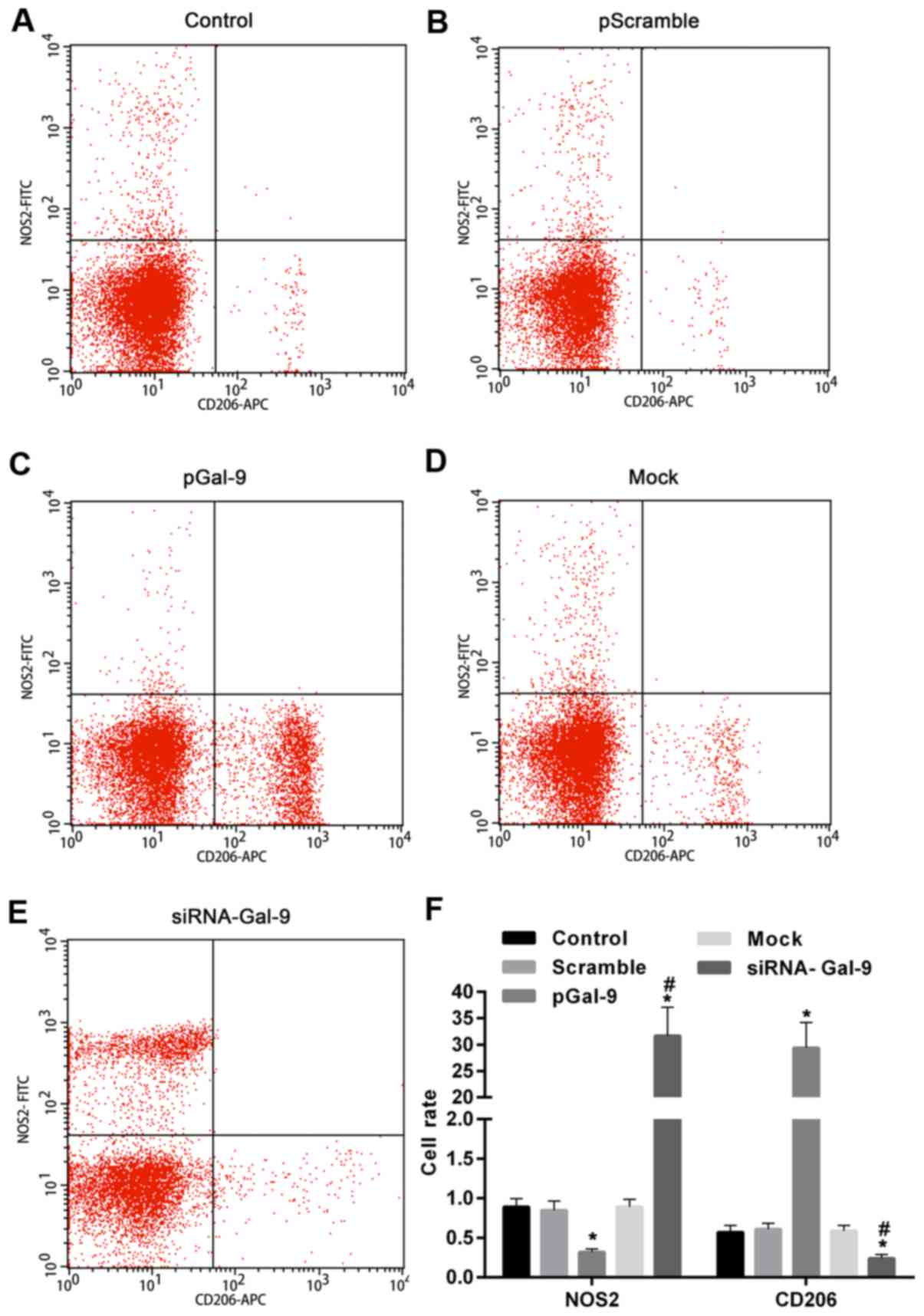
To better understand the effect of elevated or reduced expression of Gal-9 on the regulation of macrophage polarization, we detected the levels of polarization biomarkers, including NOS2 for M1-type and CD206 for M2-type. As showed in Fig. 3, flow cytometry assay revealed that NOS2 expression level was upregulated in cells transfected with siRNA-Gal-9, whereas it was downregulated in cells transfected with pGal-9, compared to cells from Control, Scramble, and Mock group. By contrast, CD206 expression level was downregulated in cells transfected with siRNA-Gal-9, whereas it was upregulated in cells transfected with pGal-9, compared to cells from Control, Scramble, and Mock group. In addition, the expression levels of NOS2 and CD206 were no apparent variation among Control, Scramble, and Mock group.
Macrophage polarization-associated genes are regulated by levels of Gal-9 in both extracellular and intracellular
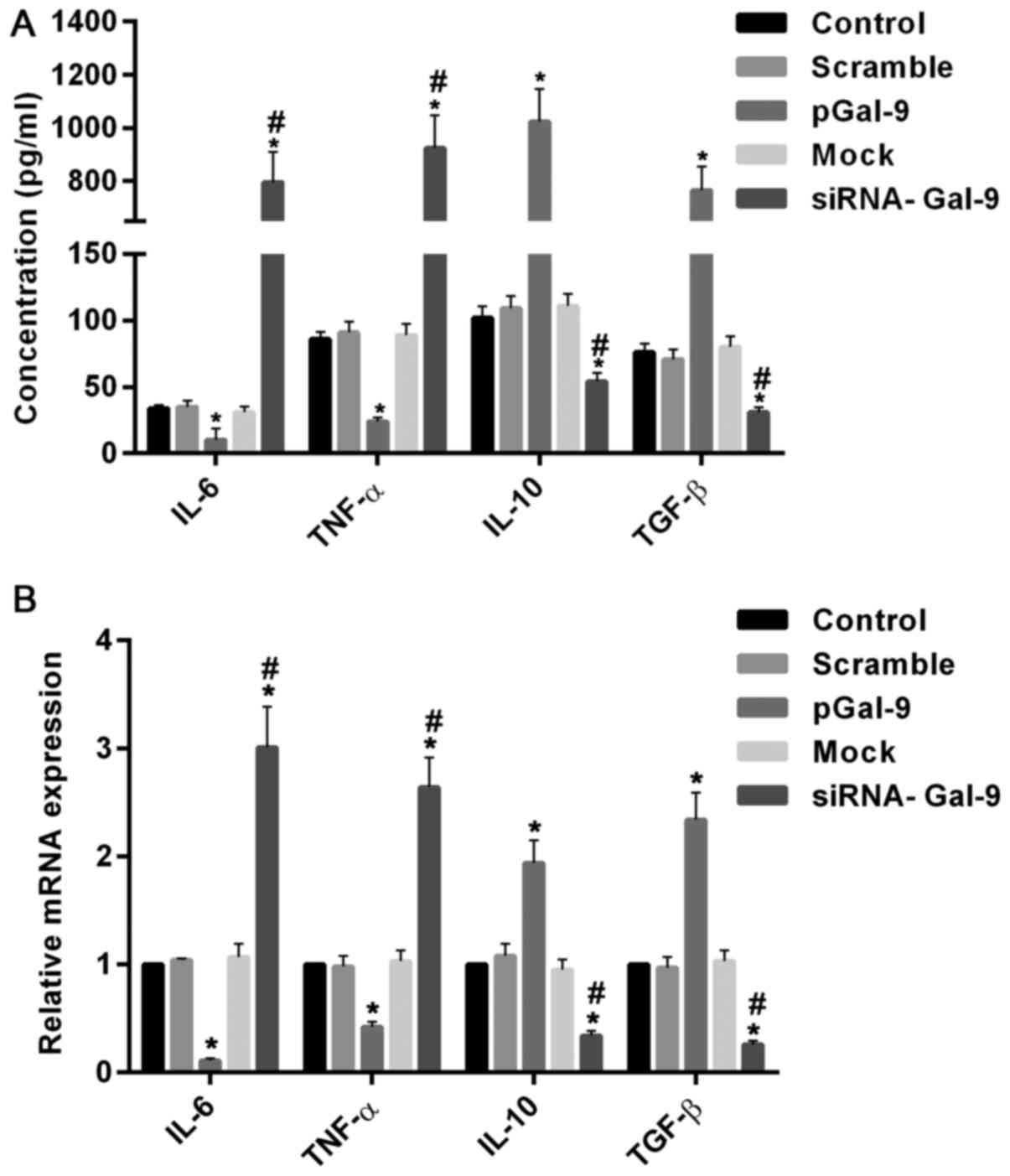
TNF-α and TGF-β were tightly related to M1-type polarization and M2-type polarization, respectively. In order to further confirm the effect of overexpression and down-regulation of Gal-9 on RAW264.7 cell polarization, we detected the protein and mRNA levels of IL-6, TNF-α, IL-10 and TGF-β using ELISA assay and real-time RT-PCR assay, respectively. Fig. 4 showed the variations of macrophage polarization-associated genes after transfection for 48 h. In cells transfected with pGal-9, IL-6 and TNF-α protein and mRNA levels were downregulated, whereas IL-10 and TGF-β protein and mRNA levels were upregulated, compared to the control groups (Control, Scramble and Mock). By contrast, in cells transfected with siRNA-Gal-9, IL-6 and TNF-α protein and mRNA levels were upregulated, whereas IL-10 and TGF-β protein and mRNA levels were downregulated, compared to the control groups. These results confirmed the observations of flow cytometry assay conducted above.
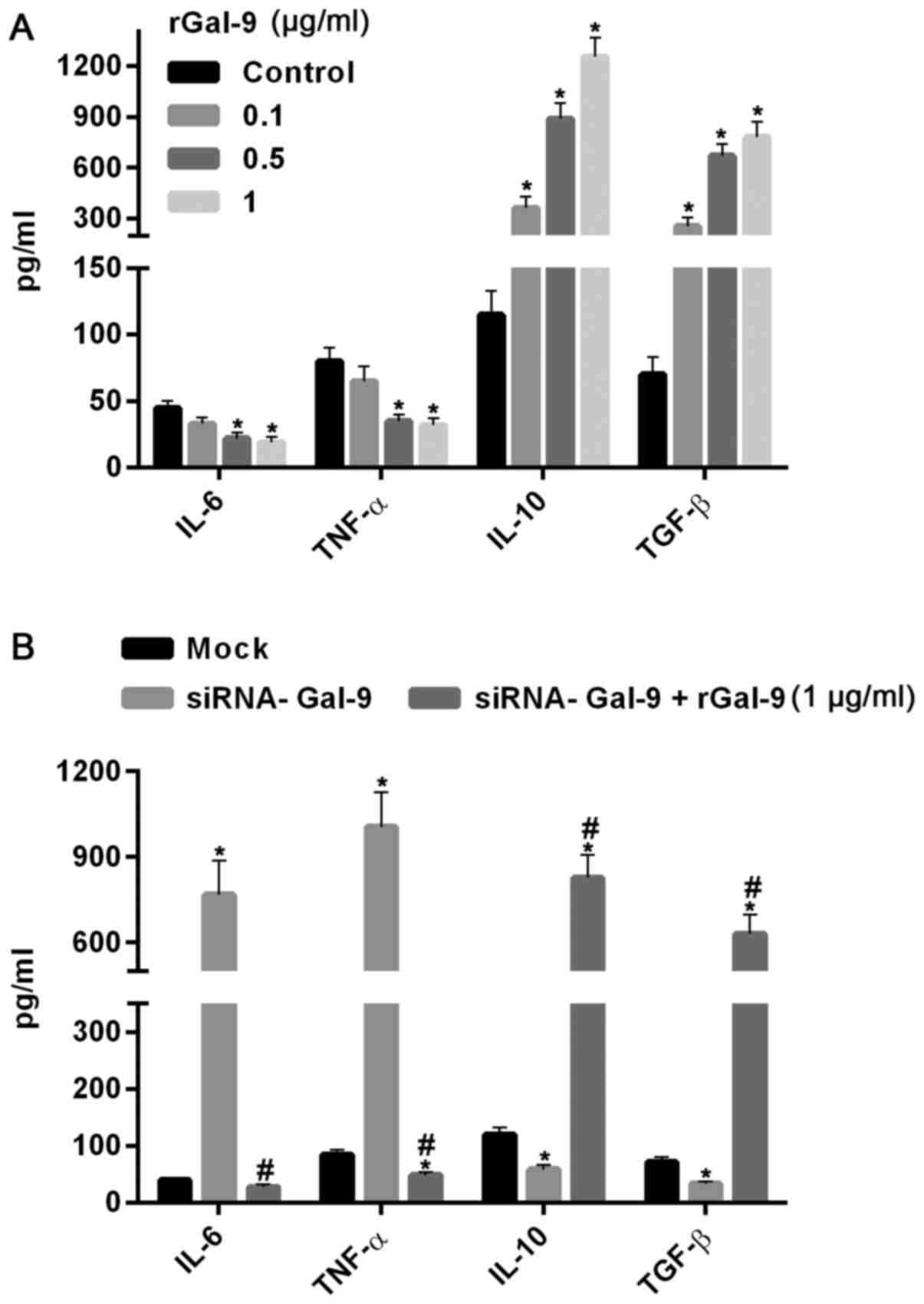
To further confirm the effect of Gal-9 on the cells, we added carrier-free recombinant Gal-9 (R&D Systems, Inc., Minneapolis, MN, USA) into the medium (19). After 24 h, the results showed that IL-6 and TNF-α were dosage-dependently decreased, whereas IL-10 and TGF-β were significantly increased with increasing levels of rGal-9 (0.1–1 µg/ml) (Fig. 5A). Moreover, addition of rGal-9 (1 µg/ml) for 24 h evidently abolished the effect of siRNA-Gal-9 on proteins expression (Fig. 5B).
Gal-9 can mediate the macrophage polarization-associated transcription factors
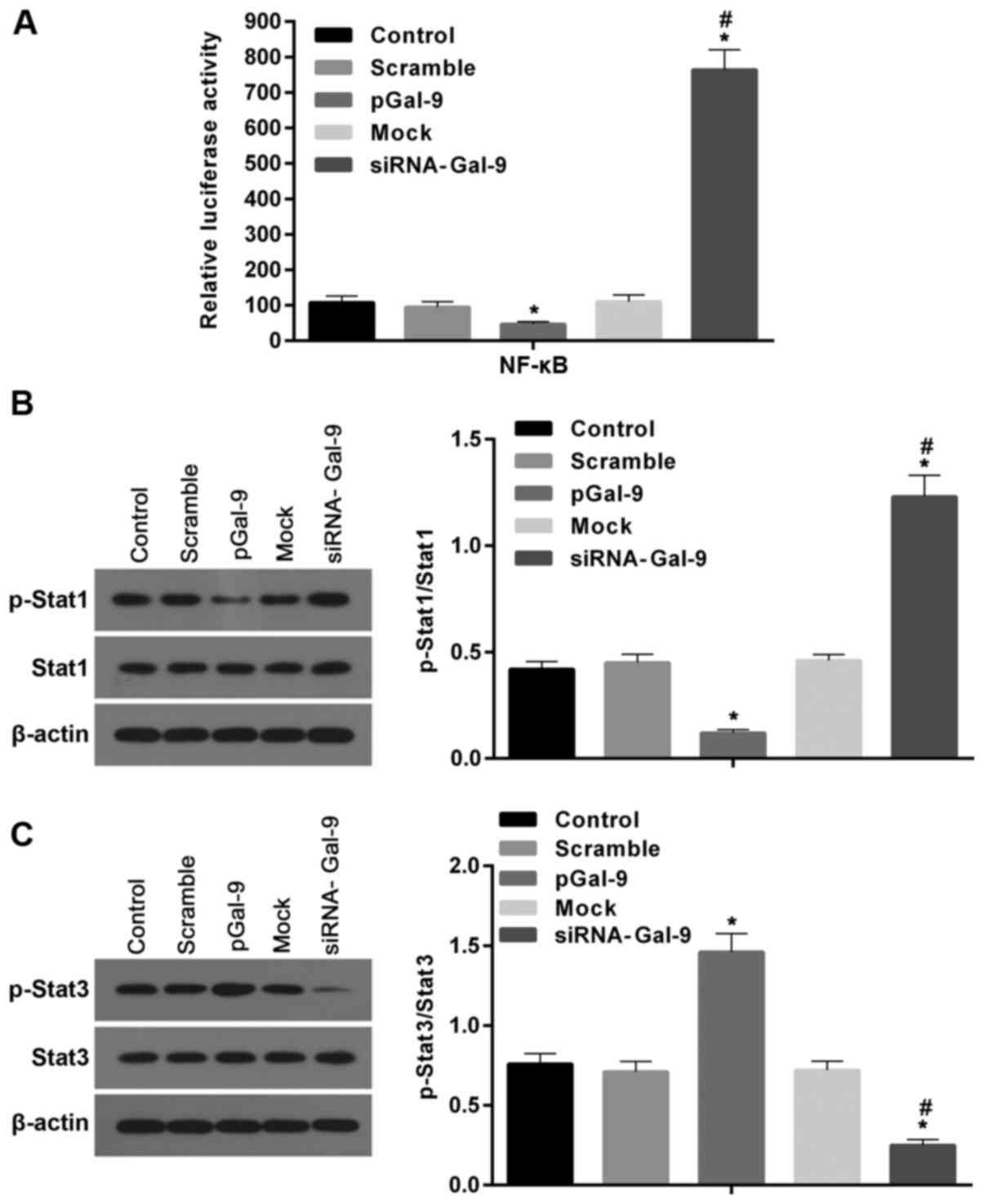
Studies revealed that transcription factors, including NF-κB, Stat1, and Stat3, are involved macrophage polarization. To detect the potential pathway through which Gal-9 mediates the macrophage polarization, we determined the expression levels of these factors after cells transfected with pGal-9 or siRNA-Gal-9. The transcription activity of NF-κB detected by dual-luciferase reporter gene assay was downregulated in cells transfected with pGal-9, whereas it was upregulated in cells transfected with siRNA-Gal-9, compared to the control groups. In addition, there were nearly equal levels among the control groups (Fig. 6A).
The western blot assay revealed that in cells transfected with pGal-9, together with the level of p-Stat1 protein, the ratio of p-Stat1 to Stat1 were reduced, though Stat1 level was no apparent changes, compared to the control (Fig. 6B). By contrast, in cells transfected with siRNA-Gal-9, both p-Stat1 protein level and the ratio of p-Stat1 to Stat1 were elevated compared to control, despite the Stat1 level was nearly equal compared with control groups (Fig. 6B). However, the Stat3 levels were altered in the opposite way compared to Stat1. Fig. 6C showed that in cells transfected with pGal-9, the level of p-Stat3 protein and the ratio of p-Stat3 to Stat3 were upregulated, though Stat3 level was no visible changes, compared to control groups. While in cells transfected with siRNA-Gal-9, p-Stat3 protein levels and the ratio of p-Stat3 to Stat3 were significantly decreased, and despite the Stat3 level was slightly increased, compared with control groups.
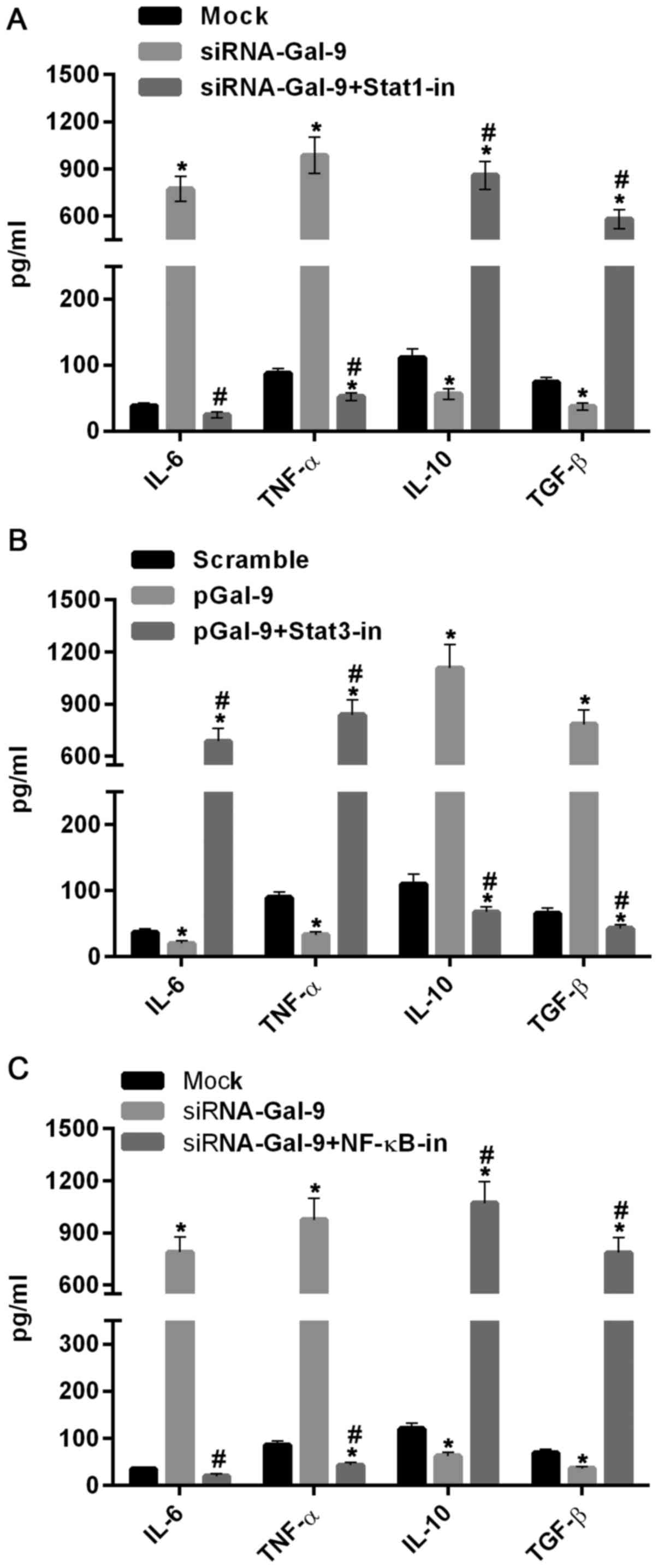
We further tested the role of Stat1, Stat3 and NF-κB in regulations of M1-type and M2-type macrophages related cytokines. As showed in Fig. 7A, the levels of IL-6 and TNF-α were elevated, whereas the levels of IL-10 and TGF-β were reduced in cells treated with siRNA-Gal-9, compared to those in cells from Mock group. By contrast, these cytokines were inversely varied in cells treated with sRNA-Gal-9 and Stat1 inhibitor Fludarabine (10 µM) (Selleck Chemicals, Houston, TX, USA). These results suggested that Stat1 was a positive transcription factor for M1-type macrophages. In Fig. 7B, the levels of IL-6 and TNF-α were decreased, while the levels of IL-10 and TGF-β were dramatically increased in cells treated with pGal-9, compared to those in cells from Scramble group. However, these cytokines were inversely changed in cells treated with pGal-9 and Stat3 inhibitor Stattic (7 µM) (Selleck Chemicals). These results indicated that Stat3 promotes the transcription of cytokines related to M2-type macrophages. Moreover, similar effects of NF-κB and its inhibitor JSH-23 (10 µM) (Selleck Chemicals) on levels of these cytokines as Stat1 was showed in Fig. 7C, which demonstrated that NF-κB can simulate expression of cytokines genes related to M1-type macrophages and suppress expression of cytokines associated with M2-type macrophages. Therefore, together with the results discussed above, this study suggested that Gal-9 induces the activation of NF-κB, Stat1 or Stat3 signaling molecules and thereby promoting the M1-type or M2-type related genes expression, resulting the shifting between M1-type and M2-type of macrophages polarization.
Discussion
Even though Gal-9 is involved in the regulation of a variety of biological processes such as cell aggregation and adhesion, chemotaxis of eosinophils, induction of thymocytes, immune T cells and induction of melanoma cell apoptosis, the role of it in macrophages is poorly understood. In our study, we had explored the expression of macrophage polarization-associated genes. Our results suggested that Gal-9 was a potential regulator for the macrophage polarization. Firstly, we successfully constructed cell models with upregulation or downregulation of Gal-9, which confirmed by protein and mRNA analysis. The further flow cytometry assay revealed that macrophage polarization indicators for M1-type, NOS2, and for type-M2, CD206, were downregulated and upregulated in cells with upregulated Gal-9. In contrast, NOS2 and CD206 were reversed in cells with knockdown of Gal-9 compared to cells with overexpression of Gal-9. These results suggested that upregulated Gal-9 may induce M2-type macrophages, while downregulated Gal-9 can simulate M1-type macrophages. In addition, the expression levels of protein and mRNA of IL-6, TNF-α, IL-10 and TGF-β confirmed our observation. Furthermore, our results showed that the regulations of Gal-9 on the macrophage polarization may be involved transcription factors NF-κB, Stat1, and Stat3. This study showed a close association of Gal-9 with the regulation of polarization in RAW264.7 cells.
Gal-9 is a member of the galactoside lectin family, which was isolated from the mouse embryo for the first time in 1997 by Wada et al (20) and is a sugar-binding protein. Tim-3 protein is an important member of the Tim family, and plays an immunomodulatory role when combined with its ligand Gal-9 (14–16). Very recently, investigation showed that Tim-3 has a steady-state regulation of macrophages polarization (18,21). Therefore, we hypothesized that Gal-9 may be the target of macrophage polarization.
Macrophage polarization is divided into two categories: One is the classic activated macrophages, also known as M1 macrophages; the other is the alternative activation of macrophages, that is, M2 macrophages (1,2). M1 macrophages and M2 macrophages can exert their biological functions by secreting different cytokines and effector molecules (4). Our results firstly showed that the levels of Gal-9 are varied not only in intracellular but also in extracellular after the cells treated with pGal-9 or siRNA-Gal-9 (Fig. 1). We then found that the cell viability was obviously decreased in cells treated with siRNA-Gal-9, while it was slightly increased in cells treated with pGal-9, compared to control (Fig. 2). These results indicated that Gal-9 is associated with the cytotoxicity or viability of macrophages. However, these findings may be associated with macrophages polarization shifting between M1 and M2 (4). NOS2 is a marker tightly connected with M1-type polarization of macrophages, where as CD206 is a widely used marker for alternatively activated (M2) macrophage (22,23). Our results showed that overexpressed Gal-9 significantly inhibited the expression of NOS2, but also increased the expression of CD206 in the flow cytometry assay (Fig. 3C-F). Ours results suggested that overexpressed Gal-9 can induce macrophage to generate M2-type macrophages, and inhibit M1-type macrophages polarization. By contrast, knockdown of Gal-9 is clearly associated with M1-type macrophages since a markedly increased NOS2 expression and reduction of CD206 expression was observed, suggesting that Gal-9 downregulation can induce M1-type macrophages polarization and inhibit M1-type macrophages polarization (Fig. 3C-F). Furthermore, M1-type macrophages are characterized by IL-12high, lL-23high, lL-10low, and efficient secretion effector molecules (e.g., ROIs) and inflammatory cytokines such as IL-6, TNF-α and IL-1β (24–26); M2-type macrophages are characterized by IL-121ow, IL-231ow, IL-10high, high expression of galactose receptors, and cannot produce inflammatory factors, NO and ROIs (27,28). In this study, we found that both IL-6 and TNF-α were suppressed at level of transcription and translation in cells with overexpressed Gal-9 in macrophages, where IL-10 and TGF-β were markedly elevated (Fig. 4), suggesting that Gal-9 overexpression in macrophages can induce M2-type macrophages. However, in cells with downregulated Gal-9 the transcriptional and translational patterns of IL-6 and TNF-α or IL-10 and TGF-β were inversely associated with that in cells with overexpressed Gal-9 (Fig. 4), suggesting that Gal-9 downregulation in macrophages can simulate M1-type macrophages. Moreover, the effect of Gal-9 on cytokines related to M1-type or M2-type macrophages was confirmed by addition of carrier-free recombinant Gal-9 in extracellular (Fig. 5). These results were consistent with the observations of decreased M1-type marker NOS2 and increased M2-type marker CD206 mentioned above. It is exciting that in another latest research, the expression levels of pro-inflammatory cytokines in M1-differentiated THP-1 cells co-cultured with hGal-9-transfected porcine kidney epithelial cells were reduced (29). Besides, this study also showed that hGal-9 has a decrease in M1-differentiated THP-1 cell cytotoxic activity-related acute immune rejection in pig-to-human xenotransplantation (29). Moreover, a similar study showed that overexpression of Tim3 or exogenous recombinant Gal9 can decrease inflammatory factor IFNγ, and elevate anti-inflammatory factor IL-10 (19). These results suggested that Gal-9 plays an important role in immunomodulatory of macrophages.
Previous investigations revealed that NF-κB is tightly connected with the cytokines exerted from M2-type macrophages (30). In this study, NF-κB activity was reduced and increased in cells with upregulated and downregulated Gal-9 (Fig. 6A), respectively, indicating that macrophages with Gal-9 overexpression was tend to generate M2-type polarization and macrophages with Gal-9 downregulation promote M1-type polarization. In addition, Statl and Stat3 are members of the signal transduction and transcription activator family (Stat), which are also involved in macrophage polarization (8,9). The Stat3 knockout mice showed that Stat3 in macrophages has an important anti-inflammatory effect, and the bactericidal ability of these mice is significantly decreased (31). Phosphorylated Stat (p-Stat)1 participates in M1-type polarization, and its expression is regulated by NF-κB (32). While p-Stat3 participates in M2-type polarization, and p-Stat3 inhibits NF-κB activation and p-Stat1 expression, thereby inhibiting macrophage M1 polarization and promoting M2-type polarization (33,34). Consistent with these reports, our results revealed that p-Stat1 as well as NF-κB activity were inhibited and elevated in cells with upregulated and downregulated Gal-9, respectively (Fig. 6B). However, p-Stat3 level was increased and decreased in cells with upregulated and downregulated Gal-9, respectively, which were inversely associated with NF-κB activity (Fig. 6C). Interestingly, no apparent variations on the levels of Stat1 and Stat3 were observed among all groups in this study. Together with previous results, we suggested that the regulation effect of Gal-9 in macrophages on the expression of TNF-α, IL-6, TGF-β, IL-10 may be mediated by the interactions between Gal-9 and NF-κB, Stat1 or Stat3. Moreover, we further confirmed this hypothesis by suppressing the activation of these signaling molecules with their specific inhibitors (Fig. 7). In a study by Jung et al (29), M2 differentiation was turned on while M1 differentiation was turned down in M1-differentiated THP-1 cells co-cultured with hGal-9-transfected porcine kidney epithelial cells via enhancing the phosphorylation levels of Akt and PI3K and the expression level of PPAR-γ. Here, the interactions between Akt/PI3K and NF-κB, Stat1 or Stat3 deserve further investigations.
In summary, our results suggested that macrophage polarization was tightly regulated with alterations of Gal-9 expression and significantly associated with Gal-9-mediated cytokines, transcription factors and regulators, including TNF-α, IL-6, TGF-β, IL-10, NF-κB, Stat1 and Stat3. Our results provide insight into the mechanism of the effect of Gal-9 overexpression or knockdown on the polarization and cytokines in macrophages.
Acknowledgements
This study was supported by The Natural Science Fund of Zhejiang Province (LQ16H150001).
References
|
Martinez FO, Sica A, Mantovani A and Locati M: Macrophage activation and polarization. Front Biosci. 13:453–461. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Mantovani A, Sozzani S, Locati M, Allavena P and Sica A: Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Wang C, Liu X, Liu Y, Zhang Q, Yao Z, Huang B, Zhang P, Li N and Cao X: Zinc finger protein 64 promotes Toll-like receptor-triggered proinflammatory and type I interferon production in macrophages by enhancing p65 subunit activation. J Biol Chem. 288:24600–24608. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Mantovani A, Sica A and Locati M: New vistas on macrophage differentiation and activation. Eur J Immunol. 37:14–16. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Macatonia SE, Hsieh CS, Murphy KM and O'Garra A: Dendritic cells and macrophages are required for Th1 development of CD4+ T cells from alpha beta TCR transgenic mice: IL-12 substitution for macrophages to stimulate IFN-gamma production is IFN-gamma-dependent. Int Immunol. 5:1119–1128. 1993. View Article : Google Scholar : PubMed/NCBI | |
|
Puddu P, Fantuzzi L, Borghi P, Varano B, Rainaldi G, Guillemard E, Malorni W, Nicaise P, Wolf SF, Belardelli F and Gessani S: IL-12 induces IFN-gamma expression and secretion in mouse peritoneal macrophages. J Immunol. 159:3490–3497. 1997.PubMed/NCBI | |
|
Bashir S, Sharma Y, Elahi A and Khan F: Macrophage polarization: The link between inflammation and related diseases. Inflamm Res. 65:1–11. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Tymoszuk P, Charoentong P, Hackl H, Spilka R, Müller-Holzner E, Trajanoski Z, Obrist P, Revillion F, Peyrat JP, Fiegl H and Doppler W: High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC Cancer. 14:2572014. View Article : Google Scholar : PubMed/NCBI | |
|
Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al: The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 11:936–944. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Tamura M, Watanabe T, Igarashi T, Takeuchi T, Kasai K and Arata Y: Crosslinking of Cys-mutated human galectin-1 to the model glycoprotein ligands asialofetuin and laminin by using a photoactivatable bifunctional reagent. Biol Pharm Bull. 37:877–882. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Vicuña L, Pardo E, Curkovic C, Döger R, Oyanadel C, Metz C, Massardo L, González A and Soza A: Galectin-8 binds to LFA-1, blocks its interaction with ICAM-1 and is counteracted by anti-Gal-8 autoantibodies isolated from lupus patients. Biol Res. 46:275–280. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Wijesundera KK, Izawa T, Tennakoon AH, Murakami H, Golbar HM, Katou-Ichikawa C, Tanaka M, Kuwamura M and Yamate J: M1- and M2-macrophage polarization in rat liver cirrhosis induced by thioacetamide (TAA), focusing on Iba1 and galectin-3. Exp Mol Pathol. 96:382–392. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Markovic B Simovic, Nikolic A, Gazdic M, Nurkovic J, Djordjevic I, Arsenijevic N, Stojkovic M, Lukic ML and Volarevic V: Pharmacological inhibition of Gal-3 in mesenchymal stem cells enhances their capacity to promote alternative activation of macrophages in dextran sulphate sodium-induced colitis. Stem Cells Int. 2016:26407462016.PubMed/NCBI | |
|
Elahi S, Niki T, Hirashima M and Horton H: Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood. 119:4192–4204. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F, He W, Yuan J, Wu K, Zhou H, Zhang W and Chen ZK: Activation of Tim-3-Galectin-9 pathway improves survival of fully allogeneic skin grafts. Transpl Immunol. 19:12–19. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Gooden MJ, Wiersma VR, Samplonius DF, Gerssen J, van Ginkel RJ, Nijman HW, Hirashima M, Niki T, Eggleton P, Helfrich W and Bremer E: Galectin-9 activates and expands human T-Helper 1 cells. PLoS One. 8:e656162013. View Article : Google Scholar : PubMed/NCBI | |
|
Chabtini L, Mfarrej B, Mounayar M, Zhu B, Batal I, Dakle PJ, Smith BD, Boenisch O, Najafian N, Akiba H, et al: TIM-3 regulates innate immune cells to induce fetomaternal tolerance. J Immunol. 190:88–96. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Z, Jiang X, Kang C, Xiao Y, Hou C, Yu J, Wang R, Xiao H, Zhou T, Wen Z, et al: Blockade of the T cell immunoglobulin and mucin domain protein 3 pathway exacerbates sepsis-induced immune deviation and immunosuppression. Clin Exp Immunol. 178:279–291. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Chen Y, Liu X, Zhang J, He X, Teng G and Yu D: Tim3/Gal9 interactions between T cells and monocytes result in an immunosuppressive feedback loop that inhibits Th1 responses in osteosarcoma patients. Int Immunopharmacol. 44:153–159. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wada J, Ota K, Kumar A, Wallner EI and Kanwar YS: Developmental regulation, expression, and apoptotic potential of galectin-9, a beta-galactoside binding lectin. J Clin Invest. 99:2452–2461. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang X, Yu J, Shi Q, Xiao Y, Wang W, Chen G, Zhao Z, Wang R, Xiao H, Hou C, et al: Tim-3 promotes intestinal homeostasis in DSS colitis by inhibiting M1 polarization of macrophages. Clin Immunol. 160:328–335. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Mock BA, Krall MM, Byrd LG, Chin H, Barton CH, Charles I, Liew FY and Blackwell J: The inducible form of nitric oxide synthase (NOS2) isolated from murine macrophages maps near the nude mutation on mouse chromosome 11. Eur J Immunogenet. 21:231–238. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Liu HF, Zhang HJ, Hu QX, Liu XY, Wang ZQ, Fan JY, Zhan M and Chen FL: Altered polarization, morphology, and impaired innate immunity germane to resident peritoneal macrophages in mice with long-term type 2 diabetes. J Biomed Biotechnol. 2012:8670232012. View Article : Google Scholar : PubMed/NCBI | |
|
Kienast K, Knorst M, Müller-Quernheim J and Ferlinz R: Modulation of IL-1beta, IL-6, IL-8, TNF-alpha, and TGF-beta secretions by alveolar macrophages under NO 2 exposure. Lung. 174:57–67. 1996.PubMed/NCBI | |
|
Gu Y, Hu X, Liu C, Qv X and Xu C: Interleukin (IL)-17 promotes macrophages to produce IL-8, IL-6 and tumour necrosis factor-alpha in aplastic anaemia. Br J Haematol. 142:109–114. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
An SJ, Pae HO, Oh GS, Choi BM, Jeong S, Jang SI, Oh H, Kwon TO, Song CE and Chung HT: Inhibition of TNF-alpha, IL-1beta and IL-6 productions and NF-kappa B activation in lipopolysaccharide-activated RAW 264.7 macrophages by catalposide, an iridoid glycoside isolated from Catalpa ovata G. Don (Bignoniaceae). Int Immunopharmacol. 2:1173–1181. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Y, Cheng S, Zhang M, Zhen L, Pang D, Zhang Q and Li Z: High-infiltration of tumor-associated macrophages predicts unfavorable clinical outcome for node-negative breast cancer. PLoS One. 8:e761472013. View Article : Google Scholar : PubMed/NCBI | |
|
Bastos KR, Alvarez JM, Marinho CR, Rizzo LV and Lima MR: Macrophages from IL-12p40-deficient mice have a bias toward the M2 activation profile. J Leukoc Biol. 71:271–278. 2002.PubMed/NCBI | |
|
Jung SH, Hwang JH, Kim SE, Kim YK, Park HC and Lee HT: Human galectin-9 on the porcine cells affects the cytotoxic activity of M1-differentiated THP-1 cells through inducing a shift in M2-differentiated THP-1 cells. Xenotransplantation. 24:2017. View Article : Google Scholar | |
|
Cao S, Zhang X, Edwards JP and Mosser DM: NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 281:26041–26050. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Ohkubo N, Suzuki Y, Aoto M, Yamanouchi J, Hirakawa S, Yasukawa M and Mitsuda N: Accelerated destruction of erythrocytes in Tie2 promoter-driven STAT3 conditional knockout mice. Life Sci. 93:380–387. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Cramer LA, Nelson SL and Klemsz MJ: Synergistic induction of the Tap-1 gene by IFN-gamma and lipopolysaccharide in macrophages is regulated by STAT1. J Immunol. 165:3190–3197. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Choi JW, Kwon MJ, Kim IH, Kim YM, Lee MK and Nam TJ: Pyropia yezoensis glycoprotein promotes the M1 to M2 macrophage phenotypic switch via the STAT3 and STAT6 transcription factors. Int J Mol Med. 38:666–674. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Tyagi A, Singh RP, Ramasamy K, Raina K, Redente EF, Dwyer-Nield LD, Radcliffe RA, Malkinson AM and Agarwal R: Growth inhibition and regression of lung tumors by silibinin: Modulation of angiogenesis by macrophage-associated cytokines and nuclear factor-kappaB and signal transducers and activators of transcription 3. Cancer Prev Res (Phila). 2:74–83. 2009. View Article : Google Scholar : PubMed/NCBI |