Exploring the regulatory role of Linc00893 in asthenozoospermia: Insights into sperm motility and SSC viability
- Authors:
- Published online on: December 8, 2023 https://doi.org/10.3892/mmr.2023.13143
- Article Number: 20
-
Copyright: © Lu et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
In recent years, the incidence of infertility has been increasing annually. According to statistics from the World Health Organization (WHO), the global infertility rate among couples has reached ~15%; therefore, infertility is a pressing issue in the field of reproductive medicine, which severely impacts social development (1). Global research has revealed that male infertility factors contribute to ~50% of infertility cases among couples, with abnormal sperm quality being one of the significant causes of male infertility (2). Reduced sperm motility, known clinically as asthenozoospermia (AS), is the most common cause of abnormal sperm quality (3). Sperm motility is crucial for sperm to penetrate the cervical mucus, reach the site of fertilization in the fallopian tubes and enter the oocyte to form a fertilized egg, thus completing fertilization. According to the WHO sixth edition semen analysis criteria (4), a diagnosis of AS can be considered when the progressive motility of sperm is <32% in two or more semen samples, when other sperm parameters are within the normal range (5). Currently, there are no definitive treatments for AS, and a number of infertile couples often rely on assisted reproductive techniques. Therefore, investigating the pathogenesis of AS will help in early intervention and improve the quality of life for patients.
In recent years, with the advancement of genomic technologies, a large number of non-coding genes have been discovered in the human genome. The transcripts of these genes are called non-coding RNAs, with RNA molecules >200 nucleotides referred to as long non-coding RNAs (lncRNAs) (6). Increasing evidence has suggested that lncRNAs serve important roles in the biological processes of sperm, including sperm formation, maturation and function (7). Zhang et al (8) reported significant differences in the expression profiles of lncRNAs between normal sperm and asthenozoospermic sperm, which may be related to sperm motility. Therefore, further investigation of these differences may improve understanding of the pathogenesis of AS. Saberiyan et al (9) demonstrated that ANO1-AS2 may affect sperm function in patients with AS and teratozoospermia by regulating the expression of the ANO1 gene. Another study discovered that long intergenic noncoding RNA 00574 (Linc00574) may affect sperm motility through the regulation of TCTE3 expression (10). These findings further confirm the significant role of lncRNAs in the pathogenesis of AS.
In our previous study, RNA sequencing and bioinformatics analysis were used to determine the interaction between lncRNAs and mRNAs in AS (11). Numerous differentially expressed lncRNAs were identified and validated through quantitative PCR (qPCR). Notably, Linc00893 (official gene name: EOLA1-DT) was revealed to be significantly downregulated in the seminal plasma of patients with AS, thus suggesting its association with the pathogenesis of AS. However, the pathological relationship between Linc00893 expression and AS, as well as its impact on cellular function and mechanisms, remains unclear. The present study aimed to explore the relationship between Linc00893 and AS, as well as to investigate its cellular function and molecular mechanisms. The findings may contribute to revealing the potential role of Linc00893 in sperm development and function, providing novel insights and approaches for the prevention and treatment of AS.
Materials and methods
Participants
For the present study, a total of 27 semen samples from patients with AS and 31 samples from a control group, which consisted of healthy individuals, were collected. The sample collection took place between November 2021 and May 2023. Sperm parameters were gathered from clinical microscopic examinations and semen analysis data. All specimens were obtained from the Hainan Women and Children's Medical Center (Haikou, China). The present study was approved by the Ethics Committee of the Hainan Women and Children's Medical Center (approval no. 2021-033) and conducted in accordance with the principles outlined in The Declaration of Helsinki. Written informed consent was obtained from all participants after they were provided with detailed information about the experiment.
The inclusion criteria for the AS group were as follows: i) Two consecutive semen analyses showing progressive motility <32%; ii) normal levels of hormones, such as follicle-stimulating hormone, testosterone and estradiol; iii) no abnormalities found during physical examination of the genitourinary system; iv) availability of complete medical records and follow-up data; v) in the absence of contraception, despite having regular sexual intercourse (at least 2–3 times per week), they have not been able to conceive within 1 year. The exclusion criteria were as follows: i) Abnormal parameters, such as fructose, acid phosphatase, liquefaction time, pH, sperm morphology and α-glucosidase in seminal plasma; ii) incomplete medical records and follow-up data; iii) refusal to provide informed consent. Inclusion criteria for the control group were as follows: i) Two consecutive routine semen analyses both indicating progressive motility >32%; ii) a normal semen analysis report within the past month and all other examinations showing normal results; iii) physiological parameters, such as age and body mass index (BMI), comparable to those of the AS group. Exclusion criteria included: i) Presence of acute or chronic diseases, or long-term medication use; ii) absence of complete medical records and follow-up data; iii) unwillingness to sign an informed consent form. There were no statistically significant differences in physiological parameters, such as age and BMI, between the AS group and the control group, as shown in Table I. Notably, immotile sperm rate was calculated as 100% minus the total sperm motility (%), and total sperm motility (%) is the sum of progressive motility and non-progressive motility.
Human SSC culture
Testicular biopsies from patients with obstructive azoospermia were employed for the generation of SSCs. Specifically, tissue was collected from a 35-year-old patient diagnosed with obstructive azoospermia in December 2021. The isolated SSCs from this biopsy were utilized in this study and in subsequent studies. Human immortalized SSCs were prepared according to a previously published protocol (12). The cells were cultured in a CO2 incubator (cat. no. 3111; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. The cells were cultured in Dulbecco's Modified Eagle's Medium/F12 (cat. no. 11320033; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (cat. no. 10099-141; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin. Passaging was performed at a ratio of 1:4, with medium changes 2–3 times per week.
Reverse transcription-qPCR (RT-qPCR)
Approximately 106 spermatogonial stem cells (SSCs) or >500 µl seminal plasma samples were collected. Subsequently, 1 ml TRIzol® (cat. no. 15596018; Invitrogen; Thermo Fisher Scientific, Inc.) was added, followed by vigorous shaking and a 5 min incubation at 4°C. Subsequently, 0.2 ml chloroform was added, and the mixture was vigorously shaken for 15 sec, followed by a 3 min incubation at 4°C. The sample was then centrifuged at 24,148.8 × g for 15 min at 4°C, and the upper aqueous phase was transferred to a new tube. An equal volume of isopropanol was added, mixed and incubated at −20°C for 20 min, followed by centrifugation at 24,148.8 × g for 15 min at 4°C to remove the supernatant. The pellet was then washed with 1 ml 75% DEPC ethanol and centrifuged at 10,732.8 × g for 5 min at 4°C, and the liquid was discarded to obtain RNA. RT was then performed according to the instructions provided with the Bestar™ qPCR RT Kit (cat. no. DBI-2220; DBI Bioscience) or the miRNA RT Reagent Kit (cat. no. 600036; Agilent Technologies, Inc.); briefly, 2 µg total RNA was used as a template and heated at 65°C for 5 min, followed by immediate cooling on ice. The reaction mixture, including gDNA Remover, 10X gDNA Remover Buffer, RNA and RNase-free water, was prepared according to the provided protocol. The mixture was incubated at 37°C for 5 min, followed by cooling on ice. Subsequently, another reaction mixture, including the aforementioned reaction mixture, 5X RT Buffer, RT Enzyme Mix and Primer Mix, was prepared. The first-strand cDNA was synthesized at 37°C for 15 min and 98°C for 5 min, and then collected for further use. Subsequently, qPCR amplification was performed. The reaction system, with a total volume of 20 µl, was prepared according to the protocol provided with 2X Taq PCR Master Mix (cat. no. DBI-2030; DBI Bioscience). The qPCR conditions were as follows: Initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 94°C for 20 sec, annealing at 58°C for 20 sec and extension at 72°C for 20 sec. Melting curve analysis was performed as follows: 94°C for 30 sec, 65°C for 30 sec and 94°C for 30 sec. Each sample was repeated three times. U6 was employed as the internal reference gene for miRNA expression, whereas GAPDH served as the internal reference gene for mRNA expression. The experimental data were analyzed using the 2−ΔΔCq method (13). The primer sequences are provided in Table II.
Vector construction and transfection
Linc00893 small interfering (si)RNA was designed and synthesized by Guangzhou Anernor Biotechnology Co., Ltd. The following three siRNAs were designed: si-Linc00893 #1, 5′-TTGACTTCATAACCAAGTTCT-3′; si-Linc00893 #2, 5′-GGCTGTTTTGAAGTCAGTATT-3′; si-Linc00893 #3, 5′-CTGTTTTGAAGTCAGTATTCA-3′; negative control (NC), 5′-TTCTCCGAACGAGTCACGTTT-3′. siRNA (50 nM) transfection was performed at 37°C using Lipofectamine® 3000 (cat. no. L3000-015; Invitrogen; Thermo Fisher Scientific, Inc.) at a ratio of 1:2 (siRNA:Lipofectamine 3000) when the cell density reached 30%. After transfection of SSCs for 6 h, the culture medium was replaced with fresh medium, and the cell culture plates were incubated at 37°C with 5% CO2 for 24 h before subsequent experiments.
The full-length human myosin heavy chain 9 (MYH9) cDNA (NM_002473.5) was amplified using PCR with PrimeSTAR HS DNA polymerase (cat. no. R010B; Takara Bio, Inc.). The primer sequences were as follows: Forward 5′-GTGGATCCGAGCTCGGTACCCGCCACCATGGCACAGCAAGCTGCCGATAAG-3′ and reverse 5′-GAAAATAAAGATATTTTATTACCGGTTTAATTAATTATTCGGCAGGTTTGGCCTCAG-3′. The PCR amplification conditions were segmented into three distinct phases: Initial denaturation for one cycle at 98°C for 5 min; followed by 30 cycles, each consisting of a denaturation step at 98°C for 10 sec, annealing at 55°C for 10 sec and extension at 72°C for 90 sec; the amplification process was finalized with one cycle of final extension at 72°C for 8 min. The amplified product was subsequently cloned into the pLVX-Puro lentiviral vector (cat. no. GV358; Shanghai GeneChem Co., Ltd.) between EcoRI and BamHI sites to generate the recombinant lentiviral expression vector pLVX-Puro-MYH9. A negative control vector, containing the vector components but lacking the gene-coding sequence, was also used for comparison. The 3rd generation lentiviral packaging system, including 4 µg pCMV–VSV-G (Shanghai GeneChem Co., Ltd), 3 µg pCMV-ΔR8.91 (Shanghai GeneChem Co., Ltd) and 1 µg pLVX-Puro-MYH9 was transfected into 293T cells (The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences) using Lipofectamine 3000 at 37°C, with the transfection process sustained for 8 h. The supernatant containing lentiviral particles was collected 48 h post-transfection, filtered through 0.45 µm filters and concentrated by ultracentrifugation at 66,125 × g for 2.5 h at 4°C. For lentiviral transduction, SSCs (30% confluence) were infected with lentiviral particles at a multiplicity of infection of 50 in the presence of 2 µg/ml polybrene. Cells were transduced for 12 h at 37°C, after which the medium was replaced with regular culture medium, and subsequent experiments were carried out 96 h after transduction. When co-transfecting cells with the MYH9 overexpression vector and si-Linc00893, the MYH9 overexpression lentivirus was transduced first, followed by transfection with the siRNA.
Cell Counting Kit 8 (CCK8) assay
The cells were seeded at a suitable density of 10,000 cells/well in a 96-well plate, with 100 µl culture medium added to each well. The plate was then placed in a cell culture incubator at 37°C and 5% CO2 for 24 h to allow cell attachment. On days 1, 2, 3, 4 and 5 after siRNA transfection, 10 µl CCK-8 reagent (cat. no. CK04; Dojindo Laboratories, Inc.) was added to the corresponding wells. The plate was gently shaken to ensure thorough mixing of the CCK-8 reagent with the culture medium. The 96-well plate was then returned to the cell culture incubator at 37°C and 5% CO2 for an additional 3 h. The absorbance of each well was measured at a wavelength of 450 nM using an ELISA reader (cat. no. ELx800; BioTek Instruments, Inc.).
EdU assay
The cells were seeded at a suitable density of 10,000 cells/well in a 96-well plate. After siRNA transfection and according to the instructions provided with the EdU assay kit (cat. no. C0078S; Beyotime Institute of Biotechnology), EdU at a final concentration of 10 nM was added to the culture medium. The cells were then cultured at 37°C for an additional 4 h to allow EdU to be taken up by the cells and incorporated into newly synthesized DNA. The culture medium was then removed, and the cells were fixed with 4% formaldehyde at room temperature for 20 min. Subsequently, 0.5% Triton X-100 was used to permeabilize the cells at room temperature for 10 min. The permeabilization solution was removed, and fluorescently labeled copper sulfate was added, followed by incubation at room temperature for 30 min. The cells were incubated with DAPI for nuclear staining at room temperature for 10 min, and then washed with PBS three times to remove excess dyes and reagents. Finally, the cells were observed, images were captured under a fluorescence microscope (DS-Fi3; Nikon Corporation) and the proliferation of cells was assessed by calculating the ratio of EdU-positive cells to total cells.
Apoptosis detection
Following siRNA transfection and vector transduction, cells were digested with 0.25% trypsin-EDTA solution and washed with PBS. Subsequently, the cell count was determined using a cell counter, the cells were centrifuged at 300 × g for 5 min at room temperature. and the supernatant was discarded. The cells were then washed twice with PBS and were suspended in 1X binding buffer at a concentration of 1×106 cells/100 µl. According to the instructions provided by the apoptosis detection kit (cat. no. 556547; BD Biosciences), 5 µl Annexin V-FITC and 5 µl propidium iodide (PI) were added to each cell sample, gently mixed and incubated at room temperature in the dark for 15 min. After incubation, 400 µl 1X binding buffer was added to each sample and gently mixed. The stained cell samples were analyzed using a flow cytometer (FACSCalibur; BD Biosciences). The fluorescence signal of Annexin V was detected using the FITC channel, and the fluorescence signal of PI was detected using the PE channel. FlowJo V10 software (FlowJo, LLC) was used to analyze the apoptotic rate of the cells.
Fluorescence in situ hybridization (FISH) detection
SSCs were cultured to 70–80% confluence, then washed with PBS and fixed with 4% formaldehyde at room temperature for 15 min. After fixation, the cells were washed with PBS again and permeabilized with 0.5% Triton X-100 for 10 min at room temperature, to enhance cell membrane permeability. Following permeabilization, the cells were washed with PBS again. The FISH detection kit was purchased from Guangzhou Ribobio Co., Ltd. (cat. no. C10910). The cells were incubated in pre-hybridization buffer at 37°C for 30 min, followed by replacement with hybridization solution containing Linc00893 FISH probe, with a concentration of 1 µM. The cells were hybridized for ≥16 h at 37°C. After hybridization, the cells were washed with wash buffer and treated with anti-fluorescence quenching reagent Fluoromount-G (cat. no. 0100-01; SouthernBiotech). Subsequently, blocking was performed using a blocking solution containing 5% bovine serum albumin (BSA; cat. no. 9048-46-8; AbMole Bioscience Inc.) at room temperature for 30 min to minimize nonspecific binding. Following treatment, the cells were washed with wash buffer again. The nuclei were stained with DAPI (cat. no. D9542; MilliporeSigma) at room temperature for 10 min, and the cells were washed with PBS once more. The cells were observed under a fluorescence microscope and images were captured. The Linc00893 FISH probe, with the following sequence: 5′-TAACACAAAGCTCTTTGCCTGCCCTCTAGCCTTCTTAACC-3′, was designed and synthesized by Huzhou Hippo Biotechnology Co., Ltd.
Dual-luciferase assay
The binding site prediction of Linc00893 and microRNA (miR)-107 was conducted using starBase (https://rnasysu.com/encori/), whereas the predicted binding site between miR-107 and MYH9 3′UTR was performed using TargetScan 7.0 (http://www.targetscan.org/). Based on the identified binding sites, corresponding wild-type and mutant sequences of Linc00893 and MYH9were designed and constructed into the psiCHECK-2 vector (Shanghai GeneChem Co., Ltd). The constructed vectors, along with the miR-107 mimic (sequence: 5′-ACUAUCGGGACAUGUUACGACGA-3′) and NC (sequence: 5′-UUUGUACUACACAAAAGUACUG-3′) designed and synthesized by Huzhou Hippo Biotechnology Co., Ltd., were co-transfected into SSCs (30% confluence) cultured in a 24-well plate using Lipofectamine 3000 transfection reagent. After 6 h of incubation at 37°C, under 5% CO2, the culture medium was replaced with complete culture medium. According to the instructions of the Dual-Glo Luciferase Assay Kit (cat. no. E1910; Promega Corporation), the cells were lysed using a lysis buffer and the supernatant was collected by centrifugation at 15,000 × g for 5 min at room temperature. Subsequently, 100 µl supernatant was mixed with 100 µl luciferase assay reagent and the firefly luciferase and Renilla luciferase activity was measured using a microplate reader (ReadMax 1200; Shanghai Shanpu Biotechnology Co., Ltd.). Normalization of the firefly luciferase results by comparison with Renilla luciferase activity.
RNA immunoprecipitation (RIP) experiment
The RIP assay kit was purchased from Guangzhou BersinBio Biotechnology Co., Ltd. (cat. no. Bes5101). Briefly, ~4×107 cells were scraped and collected in an RNase-free Eppendorf tube. After centrifugation at 4°C and 16,837.5 × g for 5 min, the supernatant was discarded and the cells were washed with PBS 1–2 times. Subsequently, the cells were lysed in 1 ml cell lysis buffer containing 10 µl protease inhibitor, 10 µl phosphatase inhibitor and 12 µl PMSF, and incubated at 4°C for 1–2 h. Finally, the cell lysate was centrifuged at 4°C and 24,148.8 × g for 15 min, and the supernatant was transferred to a new 1.5 ml Eppendorf tube and labeled. Magnetic beads (50 µl) were washed with 500 µl RIP buffer and centrifuged at 4°C and 1,509.7 × g for 1 min; this step was repeated twice. Subsequently, the beads were resuspended in 500 µl RIP buffer, and added to 5 µl Argonaute 2 (Ago2; 1:50; cat. no. ab186733; Abcam) or 5 µl IgG (1:50; cat. no. ab172730; Abcam) antibodies, followed by incubation at 4°C for 8 h. After centrifugation of the bead-antibody mixture at 4°C and 1,509.7 × g for 2 min, the supernatant was discarded, and the beads were washed once with 500 µl RIP buffer. Subsequently, 300 µl cell lysate was added to the mixture and incubated overnight at 4°C, while 30 µl lysate was taken as the input group. After incubation, the bead-antibody mixture was centrifuged at 4°C and 1,509.7 × g for 2 min, the supernatant was discarded and the beads were washed six times with 1 ml RIP buffer. Finally, the purified RNA was obtained and stored at −80°C for qPCR detection of miR-107 and Linc00893 expression.
Western blotting
For protein lysis of SSCs, RIPA buffer (cat. no. 89901; Thermo Fisher Scientific, Inc.) was used. Protein quantification was performed using the BCA (cat. no. 23225; Thermo Fisher Scientific, Inc.) method. For SDS-PAGE (cat. no. 1610184; Bio-Rad Laboratories, Inc.), glass plates were cleaned and set up in a casting stand. A 10% separating gel was prepared using a mixture of acrylamide, Tris, SDS, ammonium persulfate, TEMED and distilled water. After polymerization, a 5% stacking gel was added. Samples containing 20 µg protein and 3.5 µl marker were loaded and separated by SDS-PAGE. For protein transfer, a PVDF membrane (cat. no. IPVH00010; MilliporeSigma) was equilibrated and proteins were transferred at a constant current of 300 mA. The membrane was subsequently blocked with 5% BSA at room temperature for 30 min and incubated with MYH9 (1:1,000; cat. no. ab138498; Abcam) or GAPDH (1:1,000; cat. no. ab8245; Abcam) primary antibodies at room temperature for 1 h, followed by incubation with HRP-conjugated secondary antibodies (anti-rabbit, 1:10,000, cat. no. ab6721, Abcam; anti-mouse, 1:5,000, cat. no. ab6728, Abcam) at room temperature for 1 h. Chemiluminescence detection was performed using a mixture of reagents A and B (cat. no. 32106; Thermo Fisher Scientific, Inc.), and the signal was captured on X-ray film. The gel images were scanned and band intensities were semi-quantified using ImageJ software (v1.8.0; National Institutes of Health).
Gene Expression Omnibus (GEO) database analysis
The GSE160749 dataset from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) was extracted using the Aclbi platform (https://www.aclbi.com/static/index.html#/geo) to compare the expression difference of MYH9 between AS and control (healthy individuals) semen samples. Utilizing the GSE160749 dataset, AS and control semen samples were selected; subsequently, the present study focused on MYH9 gene expression, Utilizing the Aclbi platform for our analyses, boxplot diagrams were employed to visualize the expression differences between the two groups.
Statistical analysis
Statistical analysis and data visualization were performed using GraphPad Prism 9.0 software (Dotmatics). Differences between two groups were compared using an independent samples t-test. For datasets containing multiple groups, differences among the groups were compared using ANOVA (for parametric data) or Kruskal-Wallis test (for non-parametric data). In the event of a significant result, a suitable post hoc multiple comparisons, test such as Tukey (for parametric data) or Dunn's (for non-parametric data) was employed. The Wilcoxon rank-sum test was utilized to examine differential expression of MYH9 in the GSE160749 dataset. Pearson correlation analysis was utilized to assess the linear relationship between two variables. Normally distributed data are presented as the mean ± standard deviation, while non-normally distributed data are presented as the median and interquartile range and the Mann-Whitney U test was employed for analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Linc00893 is downregulated in AS semen samples and is positively correlated with sperm motility
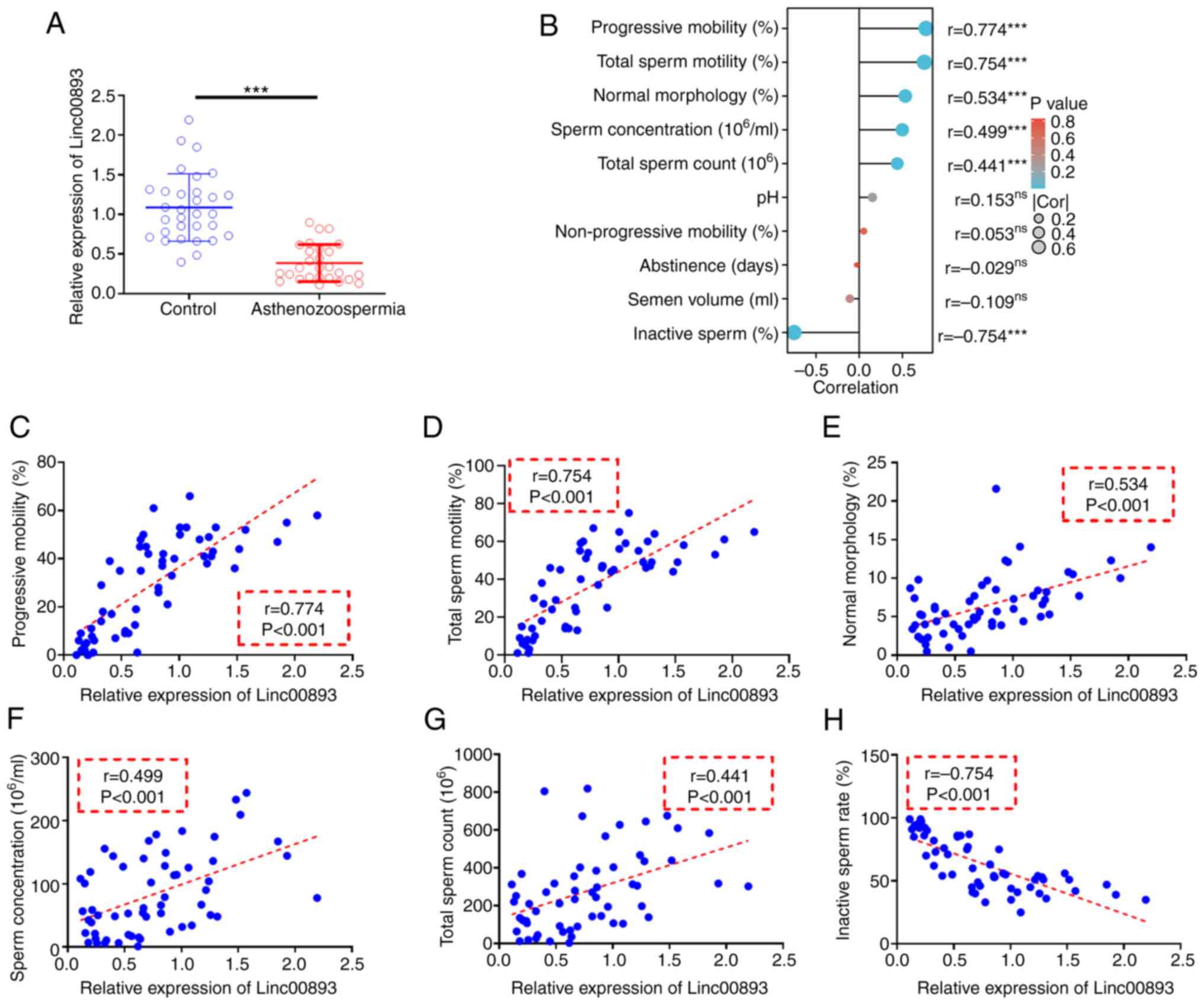
To investigate the expression of Linc00893 in AS and control semen samples, as well as its correlation with the pathology of AS, RT-qPCR was used to assess the expression levels of Linc00893 in semen. The results of RT-qPCR of clinical semen samples revealed that the expression levels of Linc00893 were significantly lower in the AS group compared with those in the control group (Fig. 1A). Pearson correlation analysis (Fig. 1B) demonstrated significant positive correlations between Linc00893 expression in semen samples and progressive mobility sperm rate (Fig. 1C), total sperm motility (Fig. 1D), normal morphology rate (Fig. 1E), sperm concentration (Fig. 1F) and total sperm count (Fig. 1G) (all r>0.3). Moreover, Linc00893 expression showed a significant negative correlation with inactive sperm rate (r<-0.3; Fig. 1H). These findings suggested that Linc00893 may be significantly downregulated in AS semen samples, and its expression is positively associated with various parameters of sperm motility, indicating its potential role in the pathology of AS, and its relevance as a biomarker for sperm motility and quality.
Knockdown of Linc00893 suppresses SSC vitality
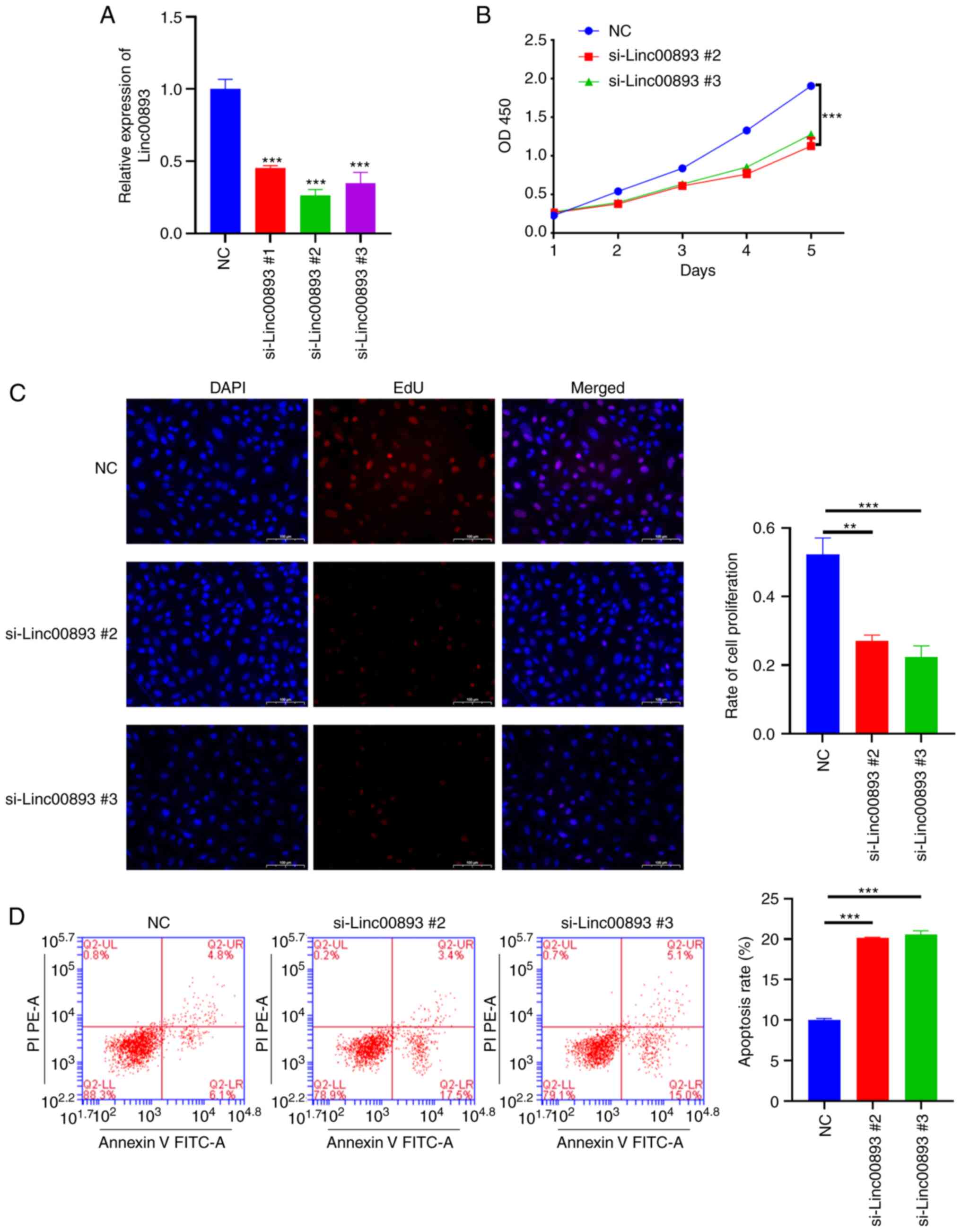
To investigate the effect of Linc00893 expression on SSCs, siRNA was used to knockdown Linc00893. To select the siRNA with the highest knockdown efficiency, three siRNA targets were designed. After transfecting SSCs with siRNAs targeting Linc00893, the results demonstrated a significant reduction in Linc00893 expression levels compared with those in the NC group, with all three siRNAs showing efficacy (Fig. 2A). Based on the knockdown efficiency, si-Linc00893 #2 and si-Linc00893 #3 were chosen for further experimentation. The CCK8 assay was performed to assess cell proliferation and revealed a significant inhibition of SSC proliferation in the si-Linc00893 groups compared with that in the NC group (Fig. 2B). In addition, EdU staining was used to assess cell proliferation, and the results demonstrated a significant suppression of SSC proliferation in the si-Linc00893 groups compared with that in the NC group (Fig. 2C). Additionally, flow cytometry was performed to examine cell apoptosis, which revealed a significant increase in the apoptotic rate (including both early and late apoptosis) of SSCs in the si-Linc00893 groups compared with that in the NC group (Fig. 2D). These results collectively indicated that knockdown of Linc00893 in SSCs may lead to a marked decrease in cell viability and proliferation, and an increase in apoptosis, highlighting the critical role of Linc00893 in maintaining SSC vitality.
Linc00893 inhibits MYH9 expression through competitive binding to miR-107
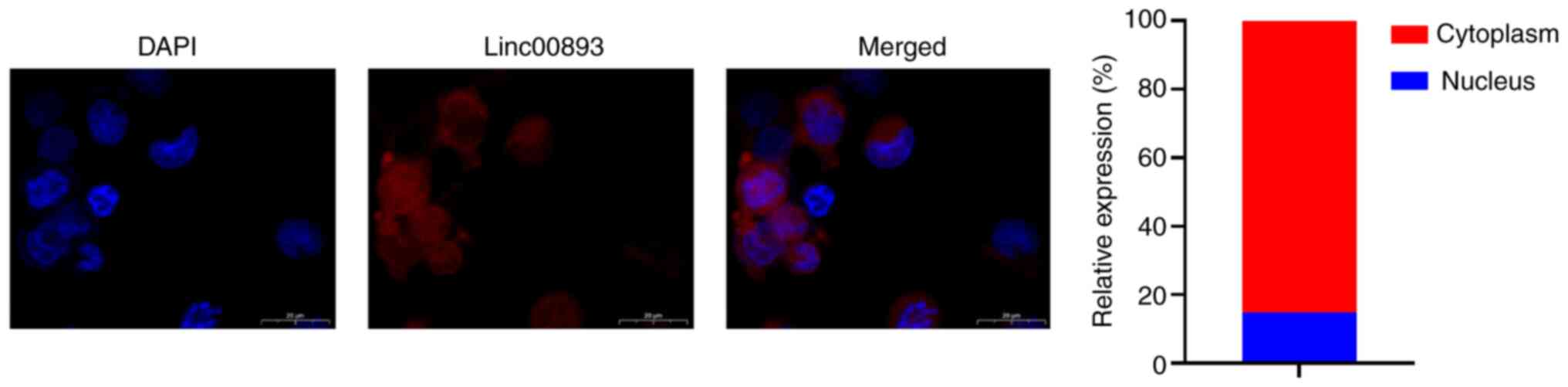
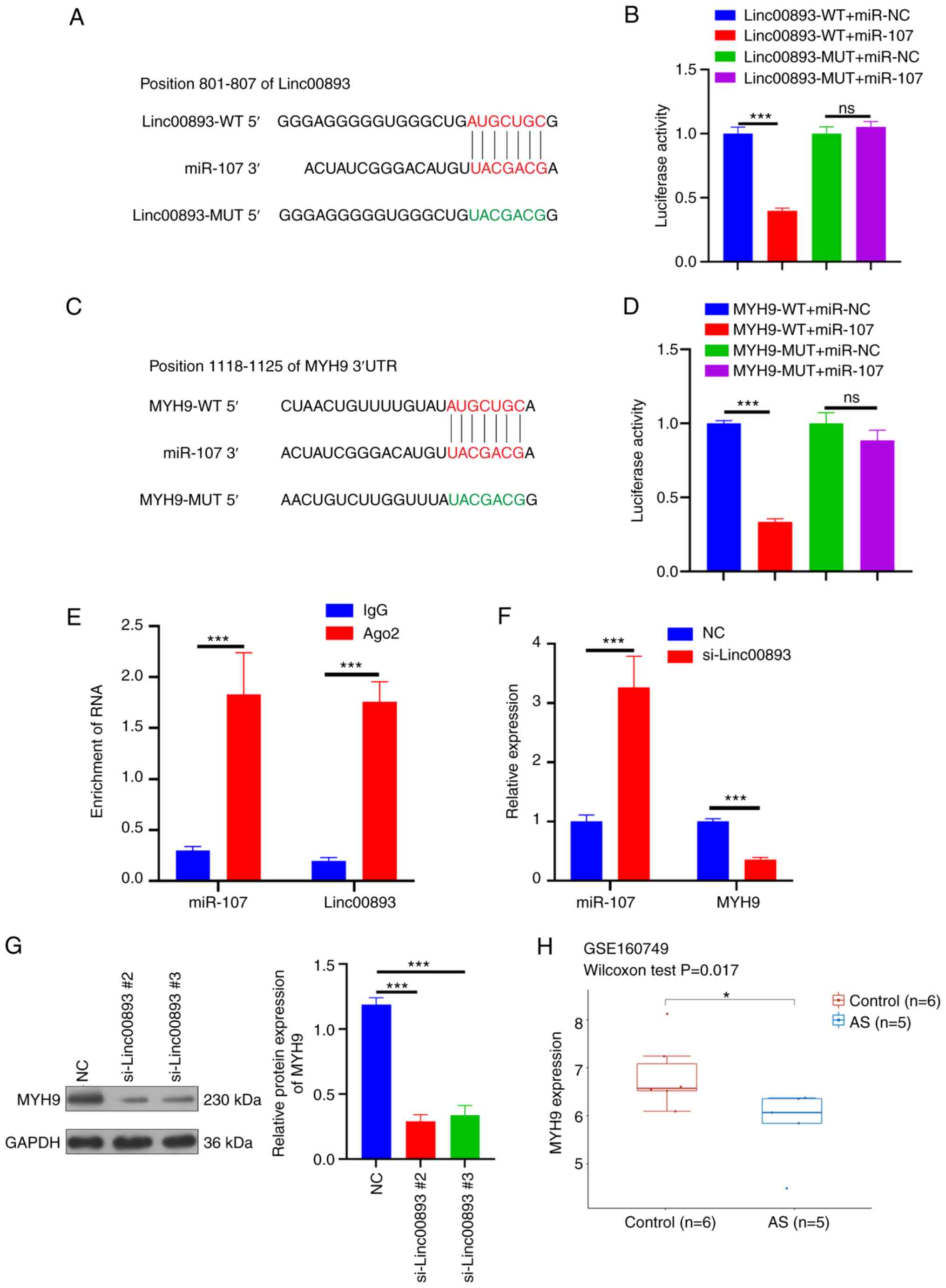
FISH was performed to determine the subcellular localization of Linc00893 in SSCs. FISH analysis of Linc00893 expression in SSCs revealed predominantly cytoplasmic localization (Fig. 3). Cytoplasmic expression of lncRNAs can potentially influence the expression of target genes through competitive endogenous RNA binding to miRNAs (14). Therefore, the miRNAs that bind to Linc00893 were predicted using the starBase database and miR-107 was selected, which has been reported to be present in sperm (15). The binding sites between Linc00893 and miR-107 were further identified, as elucidated through the starBase database (Fig. 4A). This interaction was verified using a dual-luciferase reporter assay, wherein the wild-type Linc00893 sequence was found to bind to miR-107, whereas the mutant Linc00893 sequence failed to bind (Fig. 4B). Additionally, using the TargetScan database, the target genes of miR-107 were predicted and MYH9 was selected, which has been reported to be expressed in sperm and to influence sperm function (16). The predicted binding sites between miR-107 and MYH9 were identified using the same database (Fig. 4C), and this interaction was confirmed using a dual-luciferase reporter assay, where the wild-type MYH9 sequence was found to bind to miR-107, whereas the mutant MYH9 sequence did not bind (Fig. 4D). After co-immunoprecipitation using an Ago2-specific antibody, the results of the RIP assay demonstrated significant enrichment of Linc00893 and miR-107 compared with that in the IgG control group (Fig. 4E). RT-qPCR analysis revealed that the mRNA expression levels of MYH9 were significantly decreased whereas miR-107 expression was significantly increased in the si-Linc00893 group compared with those in the NC group (Fig. 4F). Furthermore, western blot analysis revealed a significant decrease in MYH9 protein expression in the si-Linc00893 group compared with that in the NC group (P<0.001; Fig. 4G). Analysis of the GSE160749 dataset from the GEO database showed significant downregulation of MYH9 protein expression in AS samples (Fig. 4H); this finding is consistent with the downregulation trend of Linc00893 and further corroborates the mechanism where downregulated Linc00893 in AS inhibits MYH9 expression via reduced competitive binding to miR-107. These experimental outcomes suggested that Linc00893 regulates MYH9 expression by competitively binding to miR-107, thus playing a significant role in the molecular dynamics associated with AS.
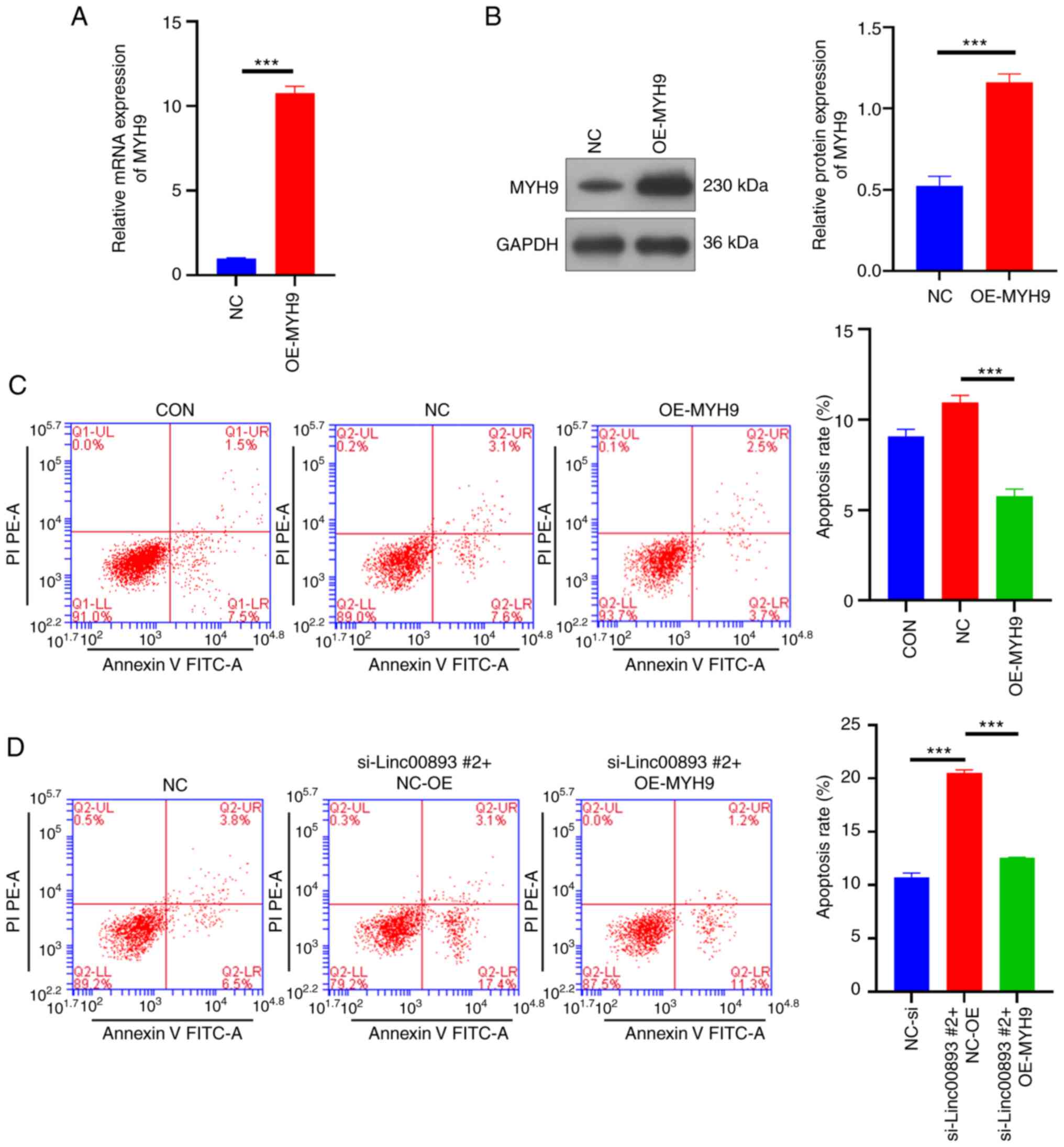
Overexpression of MYH9 inhibits SSC apoptosis and reverses apoptosis induced by Linc00893 knockdown
To confirm the impact of MYH9 on SSCs, a lentiviral vector overexpressing MYH9 and a NC vector were constructed, and were transduced into SSCs. The RT-qPCR results showed that the mRNA expression levels of MYH9 in the MYH9 overexpression (OE-MYH9) group were significantly higher compared with those in the NC group (Fig. 5A). Similarly, western blot analysis revealed that the protein expression levels of MYH9 were significantly increased in the OE-MYH9 group (P<0.001; Fig. 5B). These results suggested that MYH9 was successfully overexpressed in SSCs. The flow cytometry apoptosis assay indicated that overexpression of MYH9 inhibited SSC apoptosis (Fig. 5C), and reversed the apoptosis caused by knockdown of Linc00893 (Fig. 5D). These results indicated that overexpressing MYH9 not only reduces the apoptosis of SSCs, but also counteracts the apoptosis induced by Linc00893 knockdown, indicating the crucial role of MYH9 in modulating SSC survival and its involvement in the apoptotic pathways influenced by Linc00893.
Discussion
AS is a common cause of male infertility, characterized by reduced or lack of sperm motility (3). lncRNAs serve a critical role in male infertility, but their specific mechanisms in AS are not fully understood (7). The present study confirmed the downregulation of Linc00893 in asthenozoospermic samples, which aligns with our previous RNA sequencing results (11). This discovery suggests that Linc00893 may play an important regulatory role in the pathological process of AS. Furthermore, the expression of Linc00893 was positively correlated with progressive mobility, total sperm motility, normal morphology rate, sperm concentration and total sperm count, indicating that Linc00893 may be involved in multiple biological processes related to sperm quality. However, the sample size of 27 cases of AS is limited, precluding any subgroup analysis. AS exhibits significant heterogeneity, with varying degrees of sperm motility deficiency (17); therefore, further validation with a larger clinical sample size is necessary. The current study preliminarily indicated that Linc00893 is significantly downregulated in AS samples; given the current sample size, this preliminary conclusion can still be drawn.
SSCs are undifferentiated spermatogonia that serve as the foundation for spermatogenesis, which is responsible for the continuous production of spermatozoa (18). Additionally, SSCs undergo self-renewal throughout the lifespan of mammals, differentiating into spermatocytes and mature spermatozoa. The vitality of SSCs can influence the motility of the resulting sperm. Consequently, they serve as an in vitro model for the study of AS (13). Numerous studies have indicated that the self-renewal and proliferation of SSCs are regulated by various factors, including lncRNAs (19,20). Recent research has revealed that the expression of Linc00893 is inversely correlated with cell proliferation in various types of cancer and that it serves a pivotal role in inhibiting tumor progression (21–24). However, the relationship between Linc00893 and SSCs remains unclear. By knocking down the expression of Linc00893, the present study observed that Linc00893 knockdown inhibited SSC vitality and promoted their apoptosis. Drawing upon the present research findings from clinical samples, a positive correlation was observed between the expression of Linc00893 and the total sperm count. Furthermore, the knockdown of Linc00893 inhibited the proliferation of SSCs. Based on these observations, it may be hypothesized that reduced expression of Linc00893 can diminish male fertility by reducing sperm count and sperm vitality. Consequently, enhancing the expression of Linc00893 may be a pivotal strategy to improve the quality of male sperm.
In previous studies, the mechanism of Linc00893 has been mainly associated with its interaction with miRNAs. For example, in prostate cancer, Linc00893 interacts with miR-3173-5p, leading to the downregulation of miR-3173-5p expression. This inhibition subsequently suppresses the activation of the JAK2/STAT3 signaling pathway, thereby impeding prostate cancer progression (23). Cytoplasmic lncRNAs primarily exert their functions through interactions with miRNAs (14), which is consistent with the present finding that Linc00893 is predominantly localized in the cytoplasm. Ago2 is a central component of the RNA-induced silencing complex (RISC) (25). Within the RISC, Ago2 associates with mature miRNAs, guiding the complex to its target mRNAs, and resulting in their degradation or translational suppression (26). Consequently, the demonstration of lncRNA binding to Ago2 provides evidence for its involvement in the competitive inhibition process (27,28). The present study revealed that Linc00893 was downregulated in AS, and could reduce competitive binding to miR-107 and inhibit the expression of MYH9. MYH9, a protein encoded by the gene of the same name, is involved in cell motility and regulation of cell adhesion, and has crucial roles in diseases, such as platelet dysfunction, tumor development and hypertension (29,30). Studies have demonstrated that MYH9 is one of the key proteins involved in the binding of sperm to oviductal glycoproteins (31,32). MYH9, along with its non-muscle myosin IIB family, is essential for cytoplasmic division during male meiosis (16). However, to the best of our knowledge, there have been no reports linking MYH9 to AS. The present findings indicated that overexpression of MYH9 inhibited SSC apoptosis, suggesting a protective role for MYH9. This is consistent with the majority of literature, which has reported that MYH9 promotes cell proliferation (33–35). Therefore, it may be hypothesized that the downregulation of Linc00893, through its reduced competitive binding to miR-107, inhibits MYH9, subsequently reducing SSC viability, and contributing to the occurrence and progression of AS. The present findings may enhance understanding of the molecular mechanisms underlying AS. This knowledge is of significant importance for further elucidating the regulatory mechanisms of SSCs and developing therapeutic strategies for AS.
Although the present study has made some important discoveries, there are still limitations that should be acknowledged. Firstly, further research is needed to elucidate the precise mechanisms of Linc00893 in AS. Secondly, the present study mainly relied on in vitro experiments and lacked validation in animal models in vivo. Additionally, functional regulatory relationships between Linc00893, miR-107 and MYH9 need to be investigated through separate interventions. Finally, the clinical relevance of Linc00893 in relation to AS warrants further investigation. Large-scale clinical studies involving different populations with AS would help determine the diagnostic and prognostic value of Linc00893 in AS. Unresolved issues will be addressed in future research endeavors.
In conclusion, the present study detected the downregulation of Linc00893 in AS, and its association with sperm motility, SSC viability and MYH9 expression. Furthermore, Linc00893 may inhibit MYH9 expression through reduced competitive binding with miR-107. These discoveries provide novel insights into the pathogenesis of AS, and offer potential targets for the diagnosis and treatment of related disorders.
Acknowledgements
Not applicable.
Funding
This work was funded by the Hainan Provincial Natural Science Foundation (grant no. 822RC857) and the Key R&D Program of Hainan Province (grant no. ZDYF2023SHFZ093).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL and DX were involved in cell-related experiments and manuscript writing. LZ performed cell culture and material procurement. HR performed FISH detection. AW performed western blotting and RT-qPCR detection. JH performed dual-luciferase assay and RIP. MX performed the EdU assay and reviewed the manuscript. WL assessed cell apoptosis and reviewed the manuscript. HL and DX confirm the authenticity of all the raw data. All authors read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Affiliated Obstetrics and Gynecology Hospital of Hainan Medical College (Children's Hospital) (approval no. 2021-033) and was conducted in accordance with the principles outlined in The Declaration of Helsinki. Written informed consent was obtained from all participants (including the donor of testicular biopsies for SSC generation) after they were provided with detailed information about the experiment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Carson SA and Kallen AN: Diagnosis and management of infertility: A review. JAMA. 326:65–76. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Eisenberg ML, Esteves SC, Lamb DJ, Hotaling JM, Giwercman A, Hwang K and Cheng YS: Male infertility. Nat Rev Dis Primers. 9:492023. View Article : Google Scholar : PubMed/NCBI | |
|
Tu C, Wang W, Hu T, Lu G, Lin G and Tan YQ: Genetic underpinnings of asthenozoospermia. Best Pract Res Clin Endocrinol Metab. 34:1014722020. View Article : Google Scholar : PubMed/NCBI | |
|
Björndahl L and Brown JK; other Editorial Board Members of the WHO Laboratory Manual for the Examination and Processing of Human Semen, : The sixth edition of the WHO Laboratory manual for the examination and processing of human semen: Ensuring quality and standardization in basic examination of human ejaculates. Fertil Steril. 117:246–251. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Pan MM, Hockenberry MS, Kirby EW and Lipshultz LI: Male infertility diagnosis and treatment in the era of in vitro fertilization and intracytoplasmic sperm injection. Med Clin North Am. 102:337–347. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Herman AB, Tsitsipatis D and Gorospe M: Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 82:2252–2266. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Kyrgiafini MA, Sarafidou T and Mamuris Z: The role of long noncoding RNAs on male infertility: A systematic review and in silico analysis. Biology (Basel). 11:15102022.PubMed/NCBI | |
|
Zhang X, Zhang P, Song D, Xiong S, Zhang H, Fu J, Gao F, Chen H and Zeng X: Expression profiles and characteristics of human lncRNA in normal and asthenozoospermia sperm†. Biol Reprod. 100:982–993. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Saberiyan M, Mirfakhraie R, Gholami D, Dehdehi L and Teimori H: Investigating the regulatory function of the ANO1-AS2 on the ANO1 gene in infertile men with asthenozoospermia and terato-asthenozoospermia. Exp Mol Pathol. 117:1045282020. View Article : Google Scholar : PubMed/NCBI | |
|
Saberiyan M, Mirfakhraie R, Moghni M and Teimori H: Study of Linc00574 regulatory effect on the TCTE3 expression in sperm motility. Reprod Sci. 28:159–165. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Lu H, Xu D, Wang P, Sun W, Xue X, Hu Y, Xie C and Ma Y: RNA-sequencing and bioinformatics analysis of long noncoding RNAs and mRNAs in the asthenozoospermia. Biosci Rep. 40:BSR201940412020. View Article : Google Scholar : PubMed/NCBI | |
|
Hou J, Niu M, Liu L, Zhu Z, Wang X, Sun M, Yuan Q, Yang S, Zeng W, Liu Y, et al: Establishment and characterization of human germline stem cell line with unlimited proliferation potentials and no tumor formation. Sci Rep. 5:169222015. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar : PubMed/NCBI | |
|
Güngör BH, Tektemur A, Arkali G, Dayan Cinkara S, Acisu TC, Koca RH, Etem Önalan E, Özer Kaya S, Kizil M, Sönmez M, et al: Effect of freeze-thawing process on lipid peroxidation, miRNAs, ion channels, apoptosis and global DNA methylation in ram spermatozoa. Reprod Fertil Dev. 33:747–759. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Yang F, Wei Q, Adelstein RS and Wang PJ: Non-muscle myosin IIB is essential for cytokinesis during male meiotic cell divisions. Dev Biol. 369:356–361. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Al-Malki AH, Alrabeeah K, Mondou E, Brochu-Lafontaine V, Phillips S and Zini A: Testicular sperm aspiration (TESA) for infertile couples with severe or complete asthenozoospermia. Andrology. 5:226–231. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Lord T and Nixon B: Metabolic changes accompanying spermatogonial stem cell differentiation. Dev Cell. 52:399–411. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Hu K, Zhang J and Liang M: LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p. In Vitro Cell Dev Biol Anim. 53:277–284. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Li L, Wang M, Wang M and Wu X, Geng L, Xue Y, Wei X, Jia Y and Wu X: LncRNA analysis of mouse spermatogonial stem cells following glial cell-derived neurotrophic factor treatment. Genom Data. 5:275–278. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Li S, Zhang Y, Dong J, Li R, Yu B, Zhao W and Liu J: LINC00893 inhibits papillary thyroid cancer by suppressing AKT pathway via stabilizing PTEN. Cancer Biomark. 30:277–286. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ou X, Zhou X, Li J, Ye J, Liu H, Fang D, Cai Q, Cai S, He Y and Xu J: p53-Induced LINC00893 regulates RBFOX2 stability to suppress gastric cancer progression. Front Cell Dev Biol. 9:7964512022. View Article : Google Scholar : PubMed/NCBI | |
|
Yu C, Fan Y, Zhang Y, Liu L and Guo G: LINC00893 inhibits the progression of prostate cancer through miR-3173-5p/SOCS3/JAK2/STAT3 pathway. Cancer Cell Int. 22:2282022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu J, Jiang C, Hui H, Sun Y, Tao M, Liu Y and Qian X: Overexpressed lncRNA LINC00893 suppresses progression of colon cancer by binding with miR-146b-3p to upregulate PRSS8. J Oncol. 2022:80023182022. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Wang X, Cheng Z and Zhu Q: AGO2 and its partners: A silencing complex, a chromatin modulator, and new features. Crit Rev Biochem Mol Biol. 55:33–53. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Barbu MG, Thompson DC, Suciu N, Voinea SC, Cretoiu D and Predescu DV: The roles of MicroRNAs in male infertility. Int J Mol Sci. 22:29102021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 18:282019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao X, Su L, He X, Zhao B and Miao J: Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy. 16:70–85. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Pecci A, Ma X, Savoia A and Adelstein RS: MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene. 664:152–167. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Savoia A and Pecci A: MYH9-related disease. GeneReviews®. Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW and Amemiya A: University of Washington; Seattle, USA: (Online). | |
|
Kadam KM, D'Souza SJ, Bandivdekar AH and Natraj U: Identification and characterization of oviductal glycoprotein-binding protein partner on gametes: Epitopic similarity to non-muscle myosin IIA, MYH 9. Mol Hum Reprod. 12:275–282. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Lamy J, Nogues P, Combes-Soia L, Tsikis G, Labas V, Mermillod P, Druart X and Saint-Dizier M: Identification by proteomics of oviductal sperm-interacting proteins. Reproduction. 155:457–466. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Gao S, Wang S, Zhao Z, Zhang C, Liu Z, Ye P, Xu Z, Yi B, Jiao K, Naik GA, et al: TUBB4A interacts with MYH9 to protect the nucleus during cell migration and promotes prostate cancer via GSK3β/β-catenin signalling. Nat Commun. 13:27922022. View Article : Google Scholar : PubMed/NCBI | |
|
Hu S, Ren S, Cai Y, Liu J, Han Y, Zhao Y, Yang J, Zhou X and Wang X: Glycoprotein PTGDS promotes tumorigenesis of diffuse large B-cell lymphoma by MYH9-mediated regulation of Wnt-β-catenin-STAT3 signaling. Cell Death Differ. 29:642–656. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Lin X, Li AM, Li YH, Luo RC, Zou YJ, Liu YY, Liu C, Xie YY, Zuo S, Liu Z, et al: Silencing MYH9 blocks HBx-induced GSK3β ubiquitination and degradation to inhibit tumor stemness in hepatocellular carcinoma. Signal Transduct Target Ther. 5:132020. View Article : Google Scholar : PubMed/NCBI |