Deciphering the spatiotemporal transcriptional landscape of intestinal diseases (Review)
- Authors:
- Yajing Guo
- Chao Ren
- Yuxi He
- Yue Wu
- Xiaojun Yang
-
Affiliations: School of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan 610075, P.R. China, Graduate School, Hunan University of Traditional Chinese Medicine, Changsha, Hunan 410208, P.R. China, Department of Digestive Medicine, Chongqing City Hospital of Traditional Chinese Medicine, Chongqing 400021, P.R. China - Published online on: July 4, 2024 https://doi.org/10.3892/mmr.2024.13281
- Article Number: 157
-
Copyright: © Guo et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
This article is mentioned in:
Abstract
 |
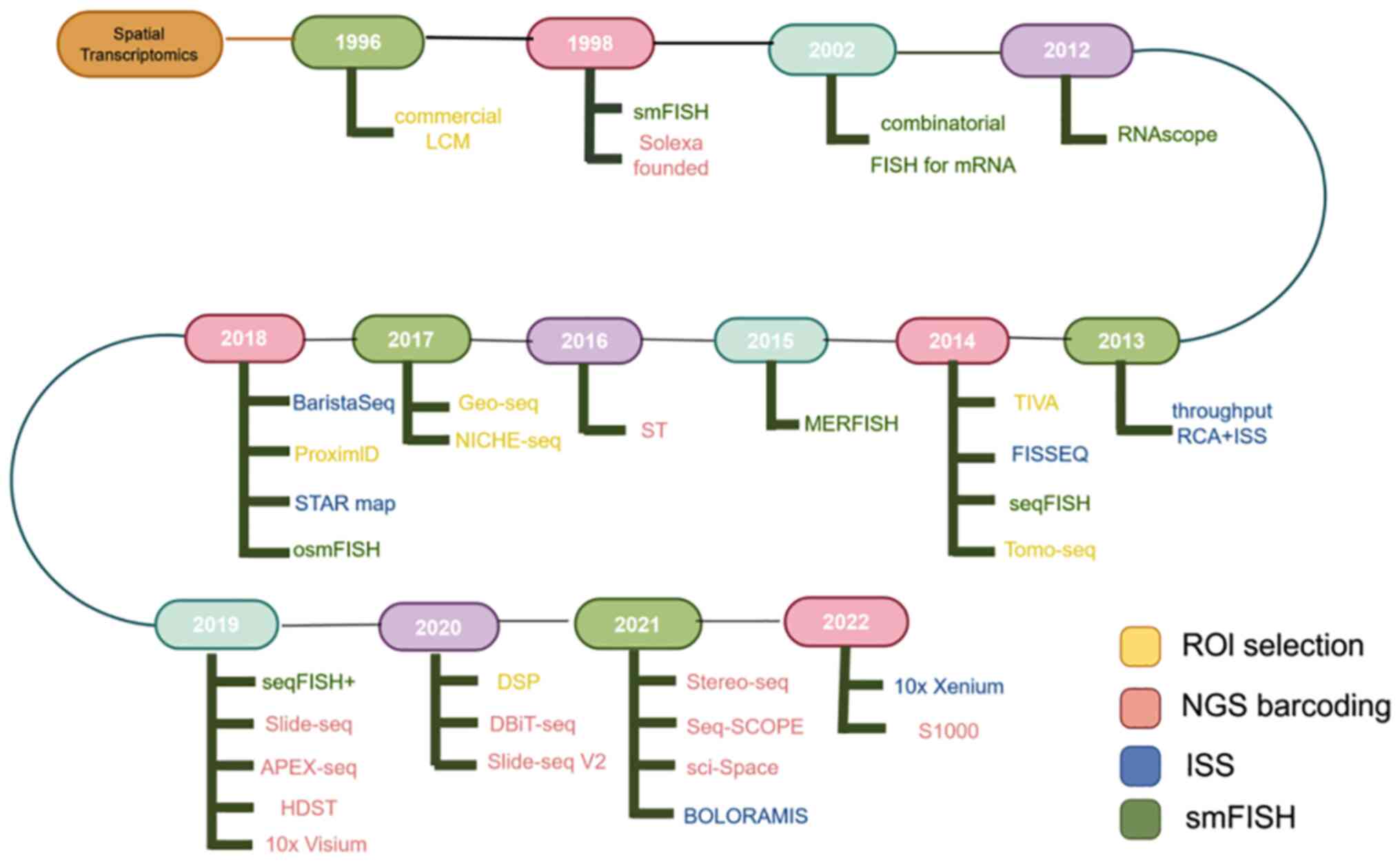 |
|
Caruso R, Lo BC and Núñez G: Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 20:411–426. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Round JL and Mazmanian SK: The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 9:313–323. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Sardinha-Silva A, Alves-Ferreira EVC and Grigg ME: Intestinal immune responses to commensal and pathogenic protozoa. Front Immunol. 13:9637232022. View Article : Google Scholar : PubMed/NCBI | |
|
Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F and Spiller RC: Functional bowel disorders. Gastroenterology. 130:1480–1491. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Mayer EA, Ryu HJ and Bhatt RR: The neurobiology of irritable bowel syndrome. Mol Psychiatry. 28:1451–1465. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Flynn S and Eisenstein S: Inflammatory bowel disease presentation and diagnosis. Surg Clin North Am. 99:1051–1062. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Bonetto S, Fagoonee S, Battaglia E, Grassini M, Saracco GM and Pellicano R: Recent advances in the treatment of irritable bowel syndrome. Pol Arch Intern Med. 131:709–715. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Saha L: Irritable bowel syndrome: Pathogenesis, diagnosis, treatment, and evidence-based medicine. World J Gastroenterol. 20:6759–6773. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Singh N and Bernstein CN: Environmental risk factors for inflammatory bowel disease. United European Gastroenterol J. 10:1047–1053. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Rosen MJ, Dhawan A and Saeed SA: Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 169:1053–1060. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang YZ and Li YY: Inflammatory bowel disease: Pathogenesis. World J Gastroenterol. 20:91–99. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Neurath MF: IL-23 in inflammatory bowel diseases and colon cancer. Cytokine Growth Factor Rev. 45:1–8. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Nadeem MS, Kumar V, Al-Abbasi FA, Kamal MA and Anwar F: Risk of colorectal cancer in inflammatory bowel diseases. Semin Cancer Biol. 64:51–60. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Brackmann S, Andersen SN, Aamodt G, Langmark F, Clausen OPF, Aadland E, Fausa O, Rydning A and Vatn MH: Relationship between clinical parameters and the colitis-colorectal cancer interval in a cohort of patients with colorectal cancer in inflammatory bowel disease. Scand J Gastroenterol. 44:46–55. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Borowitz SM: The epidemiology of inflammatory bowel disease: Clues to pathogenesis? Front Pediatr. 10:11037132023. View Article : Google Scholar : PubMed/NCBI | |
|
Kaplan GG: The global burden of IBD: From 2015 to 2025. Nat Rev Gastroenterol Hepatol. 12:720–727. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng HB, de la Morena MT and Suskind DL: The growing need to understand very early onset inflammatory bowel disease. Front Immunol. 12:6751862021. View Article : Google Scholar : PubMed/NCBI | |
|
Taylor S and Lobo AJ: Diagnosis and treatment of inflammatory bowel disease. Practitioner. 260:19–23. 2016.PubMed/NCBI | |
|
Chachu KA and Osterman MT: How to diagnose and treat IBD mimics in the refractory IBD patient who does not have IBD. Inflamm Bowel Dis. 22:1262–1274. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Parkin J and Cohen B: An overview of the immune system. Lancet. 357:1777–1789. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Kayama H, Okumura R and Takeda K: Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 38:23–48. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Groschwitz KR and Hogan SP: Intestinal barrier function: Molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 124:3–22. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Wu S, Yang L, Fu Y, Liao Z, Cai D and Liu Z: Intestinal barrier function and neurodegenerative disease. CNS Neurol Disord Drug Targets. Nov 24–2023.(Epub ahead of print). | |
|
Wu Y, Tang L, Wang B, Sun Q, Zhao P and Li W: The role of autophagy in maintaining intestinal mucosal barrier. J Cell Physiol. 234:19406–19419. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ding JH, Jin Z, Yang XX, Lou J, Shan WX, Hu YX, Du Q, Liao QS, Xie R and Xu JY: Role of gut microbiota via the gut-liver-brain axis in digestive diseases. World J Gastroenterol. 26:6141–6162. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Rutsch A, Kantsjö JB and Ronchi F: The gut-brain axis: How microbiota and host inflammasome influence brain physiology and pathology. Front Immunol. 11:6041792020. View Article : Google Scholar : PubMed/NCBI | |
|
Integrative HMP (iHMP) Research Network Consortium, . The integrative human microbiome project. Nature. 569:641–648. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Gueimonde M and Collado MC: Metagenomics and probiotics. Clin Microbiol Infect. 18 (Suppl 4):S32–S34. 2012. View Article : Google Scholar | |
|
Adak A and Khan MR: An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 76:473–493. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Patterson E, Ryan PM, Cryan JF, Dinan TG, Ross RP, Fitzgerald GF and Stanton C: Gut microbiota, obesity and diabetes. Postgrad Med J. 92:286–300. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X, Chen Y, Zhang S and Dong L: Gut microbiota-mediated immunomodulation in tumor. J Exp Clin Cancer Res. 40:2212021. View Article : Google Scholar : PubMed/NCBI | |
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 464:59–65. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Marx V: Method of the year: Spatially resolved transcriptomics. Nat Methods. 18:9–14. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al: Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 353:78–82. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Liu B, Zhao G, Lee Y, Buzdin A, Mu X, Zhao J, Chen H and Li X: Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics. 115:1106712023. View Article : Google Scholar : PubMed/NCBI | |
|
Shah S, Lubeck E, Zhou W and Cai L: In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 92:342–357. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al: Highly multiplexed subcellular RNA sequencing in situ. Science. 343:1360–1363. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al: Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 361:eaat56912018. View Article : Google Scholar : PubMed/NCBI | |
|
Asp M, Bergenstråhle J and Lundeberg J: Spatially Resolved transcriptomes-Next generation tools for tissue exploration. Bioessays. 42:e19002212020. View Article : Google Scholar : PubMed/NCBI | |
|
Shi Y, Wu X, Zhou J, Cui W, Wang J, Hu Q, Zhang S, Han L, Zhou M, Luo J, et al: Single-nucleus RNA sequencing reveals that decorin expression in the amygdala regulates perineuronal nets expression and fear conditioning response after traumatic brain injury. Adv Sci (Weinh). 9:e21041122022. View Article : Google Scholar : PubMed/NCBI | |
|
Li J, Wu C, Hu H, Qin G, Wu X, Bai F, Zhang J, Cai Y, Huang Y, Wang C, et al: Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell. 41:1152–1169.e7. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Moses L and Pachter L: Museum of spatial transcriptomics. Nat Methods. 19:534–546. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Chen KH, Boettiger AN, Moffitt JR, Wang S and Zhuang X: RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 348:aaa60902015. View Article : Google Scholar : PubMed/NCBI | |
|
Schena M, Shalon D, Davis RW and Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 270:467–470. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR and Erlander MG: Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 5:117–122. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Bhatia HS, Brunner AD, Öztürk F, Kapoor S, Rong Z, Mai H, Thielert M, Ali M, Al-Maskari R, Paetzold JC, et al: Spatial proteomics in three-dimensional intact specimens. Cell. 185:5040–5058.e19. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Alfieri CM, Mattinzoli D, Ikehata M, Cresseri D, Moroni G, Vaira V, Ferri G, Ferrero S and Messa P: Laser capture microdissection on formalin-fixed and paraffin-embedded renal transplanted biopsies: Technical perspectives for clinical practice application. Exp Mol Pathol. 116:1045162020. View Article : Google Scholar : PubMed/NCBI | |
|
Achanta S, Gorky J, Leung C, Moss A, Robbins S, Eisenman L, Chen J, Tappan S, Heal M, Farahani N, et al: A comprehensive integrated anatomical and molecular atlas of rat intrinsic cardiac nervous system. iScience. 23:1011402020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao T, Chiang ZD, Morriss JW, LaFave LM, Murray EM, Del Priore I, Meli K, Lareau CA, Nadaf NM, Li J, et al: Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 601:85–91. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Liao J, Lu X, Shao X, Zhu L and Fan X: Uncovering an organ's molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 39:43–58. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Satija R, Farrell JA, Gennert D, Schier AF and Regev A: Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 33:495–502. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Sun YM and Chen YQ: Principles and innovative technologies for decrypting noncoding RNAs: From discovery and functional prediction to clinical application. J Hematol Oncol. 13:1092020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al: Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, et al: Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 179:829–845.e20. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA and Liotta LA: Laser capture microdissection. Science. 274:998–1001. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Liew LC, Narsai R, Wang Y, Berkowitz O, Whelan J and Lewsey MG: Temporal tissue-specific regulation of transcriptomes during barley (Hordeum vulgare) seed germination. Plant J. 101:700–715. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Shaw R, Tian X and Xu J: Single-cell transcriptome analysis in plants: Advances and challenges. Mol Plant. 14:115–126. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Guo W, Hu Y, Qian J, Zhu L, Cheng J, Liao J and Fan X: Laser capture microdissection for biomedical research: Towards high-throughput, multi-omics, and single-cell resolution. J Genet Genomics. 50:641–651. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q and Hedlund E: Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun. 7:121392016. View Article : Google Scholar : PubMed/NCBI | |
|
Chen J, Suo S, Tam PP, Han JDJ, Peng G and Jing N: Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat Protoc. 12:566–580. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME and Navin NE: Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell. 172:205–217.e12. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J and van Oudenaarden A: Genome-wide RNA Tomography in the zebrafish embryo. Cell. 159:662–675. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C and Zhuang X: Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 362:eaau53242018. View Article : Google Scholar : PubMed/NCBI | |
|
Xia C, Fan J, Emanuel G, Hao J and Zhuang X: Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci USA. 116:19490–19499. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Femino AM, Fay FS, Fogarty K and Singer RH: Visualization of single RNA transcripts in situ. Science. 280:585–590. 1998. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ and Luo Y: RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 14:22–29. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M and Cai L: Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 11:360–361. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F and Macosko EZ: Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 363:1463–1467. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY and Ting AY: Atlas of subcellular RNA localization revealed by APEX-seq. Cell. 178:473–490.e26. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, et al: High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 16:987–990. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Srivatsan SR, Regier MC, Barkan E, Franks JM, Packer JS, Grosjean P, Duran M, Saxton S, Ladd JJ, Spielmann M, et al: Embryo-scale, single-cell spatial transcriptomics. Science. 373:111–117. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Yang M, Deng Y, Su G, Enninful A, Guo CC, Tebaldi T, Zhang D, Kim D, Bai Z, et al: High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 183:1665–1681.e18. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Su G, Qin X, Enninful A, Bai Z, Deng Y, Liu Y and Fan R: Spatial multi-omics sequencing for fixed tissue via DBiT-seq. STAR Protoc. 2:1005322021. View Article : Google Scholar : PubMed/NCBI | |
|
Dixon EE, Wu H, Sulvarán-Guel E, Guo J and Humphreys BD: Spatially resolved transcriptomics and the kidney: Many opportunities. Kidney Int. 102:482–491. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, Qiu X, Yang J, Xu J, Hao S, et al: Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 185:1777–1792.e21. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Cho CS, Xi J, Si Y, Park SR, Hsu JE, Kim M, Jun G, Kang HM and Lee JH: Microscopic examination of spatial transcriptome using Seq-Scope. Cell. 184:3559–3572.e22. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Levsky JM, Shenoy SM, Pezo RC and Singer RH: Single-cell gene expression profiling. Science. 297:836–840. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Lubeck E and Cai L: Single-cell systems biology by super- resolution imaging and combinatorial labeling. Nat Methods. 9:743–748. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C and Nilsson M: In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 10:857–860. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al: Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 10:442–458. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, Gustafsson A, Bernhardsson AK, Zhang C, Bohlin K and Brodin P: Stereotypic immune system development in newborn children. Cell. 174:1277–1292.e14. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Soderholm AT and Pedicord VA: Intestinal epithelial cells: At the interface of the microbiota and mucosal immunity. Immunology. 158:267–280. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Schreurs RRCE, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, Schaltenberg N, Steinert FL, van Rijn JM, Drewniak A, et al: Human fetal TNF-α-cytokine-producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. 50:462–476.e8. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Olsen TK and Baryawno N: Introduction to single-cell RNA sequencing. Curr Protoc Mol Biol. 122:e572018. View Article : Google Scholar : PubMed/NCBI | |
|
Rodaway A and Patient R: Mesendoderm. An ancient germ layer? Cell. 105:169–172. 2001.PubMed/NCBI | |
|
Zorn AM and Wells JM: Molecular basis of vertebrate endoderm development. Int Rev Cytol. 259:49–111. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Zorn AM and Wells JM: Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 25:221–251. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Kimelman D and Griffin KJ: Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. 10:350–356. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Spence JR, Lauf R and Shroyer NF: Vertebrate intestinal endoderm development. Dev Dyn. 240:501–520. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH and Hogan BL: Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 134:2521–2531. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Sherwood RI, Chen TYA and Melton DA: Transcriptional dynamics of endodermal organ formation. Dev Dyn. 238:29–42. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Walton KD, Whidden M, Kolterud Å, Shoffner SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S and Gumucio DL: Villification in the mouse: Bmp signals control intestinal villus patterning. Development. 143:427–436. 2016.PubMed/NCBI | |
|
Grey RD: Morphogenesis of intestinal villi. I. Scanning electron microscopy of the duodenal epithelium of the developing chick embryo. J Morphol. 137:193–213. 1972. View Article : Google Scholar : PubMed/NCBI | |
|
Fawkner-Corbett D, Antanaviciute A, Parikh K, Jagielowicz M, Gerós AS, Gupta T, Ashley N, Khamis D, Fowler D, Morrissey E, et al: Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell. 184:810–826.e23. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, et al: Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 178:1493–1508.e20. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, et al: Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 567:49–55. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al: Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 178:714–730.e22. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Fenderico N, van Scherpenzeel RC, Goldflam M, Proverbio D, Jordens I, Kralj T, Stryeck S, Bass TZ, Hermans G, Ullman C, et al: Anti-LRP5/6 VHHs promote differentiation of Wnt-hypersensitive intestinal stem cells. Nat Commun. 10:3652019. View Article : Google Scholar : PubMed/NCBI | |
|
Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al: Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 175:372–386.e17. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Powell DW, Pinchuk IV, Saada JI, Chen X and Mifflin RC: Mesenchymal cells of the intestinal lamina propria. Annu Rev Physiol. 73:213–237. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S, et al: Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell. 26:391–402.e5. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Degirmenci B, Valenta T, Dimitrieva S, Hausmann G and Basler K: GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 558:449–453. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al: Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 14:1099–1104. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
van de Pavert SA and Mebius RE: New insights into the development of lymphoid tissues. Nat Rev Immunol. 10:664–674. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Scheich S, Chen J, Liu J, Schnütgen F, Enssle JC, Ceribelli M, Thomas CJ, Choi J, Morris V, Hsiao T, et al: Targeting N-linked glycosylation for the therapy of aggressive lymphomas. Cancer Discov. 13:1862–1883. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Guan Q: A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res. 2019:72472382019. View Article : Google Scholar : PubMed/NCBI | |
|
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al: mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 6:377–382. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Yu X, Abbas-Aghababazadeh F, Chen YA and Fridley BL: Statistical and bioinformatics analysis of data from bulk and single-cell RNA sequencing experiments. Methods Mol Biol. 2194:143–175. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Nath A and Bild AH: Leveraging single-cell approaches in cancer precision medicine. Trends Cancer. 7:359–372. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Czarnewski P, Parigi SM, Sorini C, Diaz OE, Das S, Gagliani N and Villablanca EJ: Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun. 10:28922019. View Article : Google Scholar : PubMed/NCBI | |
|
Parigi SM, Larsson L, Das S, Ramirez Flores RO, Frede A, Tripathi KP, Diaz OE, Selin K, Morales RA, Luo X, et al: The spatial transcriptomic landscape of the healing mouse intestine following damage. Nat Commun. 13:8282022. View Article : Google Scholar : PubMed/NCBI | |
|
Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al: Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 569:121–125. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM and Smith PD: Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 115:66–75. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Martinez FO and Gordon S: The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 6:132014. View Article : Google Scholar : PubMed/NCBI | |
|
Garrido-Trigo A, Corraliza AM, Veny M, Dotti I, Melón-Ardanaz E, Rill A, Crowell HL, Corbí Á, Gudiño V, Esteller M, et al: Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat Commun. 14:45062023. View Article : Google Scholar : PubMed/NCBI | |
|
Schoeler M and Caesar R: Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. 20:461–472. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, et al: The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 81:e00036–17. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Human Microbiome Project Consortium, . Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al: Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 569:655–662. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang J, Lu H, Yin S, Ji J, Zhou L and Zheng S: Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 12:1022020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang SL, Cheng LS, Zhang ZY, Sun HT and Li JJ: Untangling determinants of gut microbiota and tumor immunologic status through a multi-omics approach in colorectal cancer. Pharmacol Res. 188:1066332023. View Article : Google Scholar : PubMed/NCBI | |
|
Pereira FC and Berry D: Microbial nutrient niches in the gut. Environ Microbiol. 19:1366–1378. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Kim HJ, Boedicker JQ, Choi JW and Ismagilov RF: Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 105:18188–18193. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Rausch P, Basic M, Batra A, Bischoff SC, Blaut M, Clavel T, Gläsner J, Gopalakrishnan S, Grassl GA, Günther C, et al: Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int J Med Microbiol. 306:343–355. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, Young VB and Sun D: Spatial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere. 4:e00126–19. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Sheth RU, Li M, Jiang W, Sims PA, Leong KW and Wang HH: Spatial metagenomic characterization of microbial biogeography in the gut. Nat Biotechnol. 37:877–883. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Choi CH and Chang SK: Role of small intestinal bacterial overgrowth in functional gastrointestinal disorders. J Neurogastroenterol Motil. 22:3–5. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ and Hardcastle JD: Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 29:1035–1041. 1988. View Article : Google Scholar : PubMed/NCBI | |
|
Macfarlane GT, Gibson GR and Cummings JH: Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol. 72:57–64. 1992. View Article : Google Scholar : PubMed/NCBI | |
|
Birchenough GM, Johansson ME, Gustafsson JK, Bergström JH and Hansson GC: New developments in goblet cell mucus secretion and function. Mucosal Immunol. 8:712–719. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Kotarsky K, Sitnik KM, Stenstad H, Kotarsky H, Schmidtchen A, Koslowski M, Wehkamp J and Agace WW: A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 3:40–48. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
McCallum G and Tropini C: The gut microbiota and its biogeography. Nat Rev Microbiol. 22:105–118. 2024. View Article : Google Scholar : PubMed/NCBI | |
|
Cremer J, Arnoldini M and Hwa T: Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc Natl Acad Sci USA. 114:6438–6443. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Cremer J, Segota I, Yang CY, Arnoldini M, Sauls JT, Zhang Z, Gutierrez E, Groisman A and Hwa T: Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc Natl Acad Sci USA. 113:11414–11419. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Nagara Y, Takada T, Nagata Y, Kado S and Kushiro A: Microscale spatial analysis provides evidence for adhesive monopolization of dietary nutrients by specific intestinal bacteria. PLoS One. 12:e01754972017. View Article : Google Scholar : PubMed/NCBI | |
|
Walker AW, Duncan SH, Harmsen HJ, Holtrop G, Welling GW and Flint HJ: The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 10:3275–3283. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Leitch ECMW, Walker AW, Duncan SH, Holtrop G and Flint HJ: Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 9:667–679. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, de'Angelis N, Rabot S, Canoui-Poitrine F, Mestivier D, et al: Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci USA. 116:24285–24295. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al: Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505:559–563. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Nguyen J, Pepin DM and Tropini C: Cause or effect? The spatial organization of pathogens and the gut microbiota in disease. Microbes Infect. 23:1048152021. View Article : Google Scholar : PubMed/NCBI | |
|
Borghini R, Donato G, Alvaro D and Picarelli A: New insights in IBS-like disorders: Pandora's box has been opened; a review. Gastroenterol Hepatol Bed Bench. 10:79–89. 2017.PubMed/NCBI | |
|
Ng QX, Soh AYS, Loke W, Lim DY and Yeo WS: The role of inflammation in irritable bowel syndrome (IBS). J Inflamm Res. 11:345–349. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Qiao LY and Tiwari N: Spinal neuron-glia-immune interaction in cross-organ sensitization. Am J Physiol Gastrointest Liver Physiol. 319:G748–G760. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, et al: Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain. 142:1215–1226. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Tavares-Ferreira D, Shiers S, Ray PR, Wangzhou A, Jeevakumar V, Sankaranarayanan I, Cervantes AM, Reese JC, Chamessian A, Copits BA, et al: Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci Transl Med. 14:eabj81862022. View Article : Google Scholar : PubMed/NCBI | |
|
Bonaz BL and Bernstein CN: Brain-gut interactions in inflammatory bowel disease. Gastroenterology. 144:36–49. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Tsigos C and Chrousos GP: Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J Psychosom Res. 53:865–871. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Kitaoka S: Inflammation in the brain and periphery found in animal models of depression and its behavioral relevance. J Pharmacol Sci. 148:262–266. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
De Bellis MD, Casey BJ, Dahl RE, Birmaher B, Williamson DE, Thomas KM, Axelson DA, Frustaci K, Boring AM, Hall J and Ryan ND: A pilot study of amygdala volumes in pediatric generalized anxiety disorder. Biol Psychiatry. 48:51–57. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Schienle A, Ebner F and Schäfer A: Localized gray matter volume abnormalities in generalized anxiety disorder. Eur Arch Psychiatry Clin Neurosci. 261:303–307. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Qiao J, Li A, Cao C, Wang Z, Sun J and Xu G: Aberrant functional network connectivity as a biomarker of generalized anxiety disorder. Front Hum Neurosci. 11:6262017. View Article : Google Scholar : PubMed/NCBI | |
|
Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C and Zhuang X: High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci USA. 113:14456–14461. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou X, Lu Y, Zhao F, Dong J, Ma W, Zhong S, Wang M, Wang B, Zhao Y, Shi Y, et al: Deciphering the spatial-temporal transcriptional landscape of human hypothalamus development. Cell Stem Cell. 29:328–343.e5. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al: A mesoscale connectome of the mouse brain. Nature. 508:207–214. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, Wauters E, Pomella V, Verbandt S, Busschaert P, et al: A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 30:745–762. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Galeano Niño JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, Futran N, Houlton J, Sather C, Sicinska E, et al: Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 611:810–817. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Ozato Y, Kojima Y, Kobayashi Y, Hisamatsu Y, Toshima T, Yonemura Y, Masuda T, Kagawa K, Goto Y, Utou M, et al: Spatial and single-cell transcriptomics decipher the cellular environment containing HLA-G+ cancer cells and SPP1+ macrophages in colorectal cancer. Cell Rep. 42:1119292023. View Article : Google Scholar : PubMed/NCBI | |
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao W, Dai S, Yue L, Xu F, Gu J, Dai X and Qian X: Emerging mechanisms progress of colorectal cancer liver metastasis. Front Endocrinol (Lausanne). 13:10815852022. View Article : Google Scholar : PubMed/NCBI | |
|
Brouwer NP, Webbink L, Haddad TS, Rutgers N, van Vliet S, Wood CS, Jansen PW, Lafarge MW; imCMS Consortium; de Wilt JH, ; et al: Transcriptomics and proteomics reveal distinct biology for lymph node metastases and tumour deposits in colorectal cancer. J Pathol. 261:401–412. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Kobayashi H, Gieniec KA, Lannagan TRM, Wang T, Asai N, Mizutani Y, Iida T, Ando R, Thomas EM, Sakai A, et al: The origin and contribution of cancer-associated fibroblasts in colorectal carcinogenesis. Gastroenterology. 162:890–906. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Peng Z, Ye M, Ding H, Feng Z and Hu K: Spatial transcriptomics atlas reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment components in colorectal cancer. J Transl Med. 20:3022022. View Article : Google Scholar : PubMed/NCBI | |
|
Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, Ding X, Bao R, Hong L, Jia W, et al: Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat Commun. 13:17422022. View Article : Google Scholar : PubMed/NCBI | |
|
Valdeolivas A, Amberg B, Giroud N, Richardson M, Gálvez EJC, Badillo S, Julien-Laferrière A, Túrós D, Voith von Voithenberg L, Wells I, et al: Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics. NPJ Precis Oncol. 8:102024. View Article : Google Scholar : PubMed/NCBI | |
|
Roelands J, van der Ploeg M, Ijsselsteijn ME, Dang H, Boonstra JJ, Hardwick JCH, Hawinkels LJAC, Morreau H and de Miranda NFCC: Transcriptomic and immunophenotypic profiling reveals molecular and immunological hallmarks of colorectal cancer tumourigenesis. Gut. 72:1326–1339. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Zwing N, Failmezger H, Ooi CH, Hibar DP, Cañamero M, Gomes B, Gaire F and Korski K: Analysis of spatial organization of suppressive myeloid cells and effector T cells in colorectal cancer-A potential tool for discovering prognostic biomarkers in clinical research. Front Immunol. 11:5502502020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu HT, Chen SY, Peng LL, Zhong L, Zhou L, Liao SQ, Chen ZJ, Wang QL, He S and Zhou ZH: Spatially resolved transcriptomics revealed local invasion-related genes in colorectal cancer. Front Oncol. 13:10890902023. View Article : Google Scholar : PubMed/NCBI | |
|
Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J and Bouvier AM: Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 244:254–259. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, et al: Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 51:1113–1122. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F, Long J, Li L, Wu ZX, Da TT, Wang XQ, Huang C, Jiang YH, Yao XQ, Ma HQ, et al: Single-cell and spatial transcriptome analysis reveals the cellular heterogeneity of liver metastatic colorectal cancer. Sci Adv. 9:eadf54642023. View Article : Google Scholar : PubMed/NCBI | |
|
Garbarino O, Lambroia L, Basso G, Marrella V, Franceschini B, Soldani C, Pasqualini F, Giuliano D, Costa G, Peano C, et al: Spatial resolution of cellular senescence dynamics in human colorectal liver metastasis. Aging Cell. 22:e138532023. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng LK, O'Grady G, Du P, Egbuji JU, Windsor JA and Pullan AJ: Gastrointestinal system. Wiley Interdiscip Rev Syst Biol Med. 2:65–79. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Peterson LW and Artis D: Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat Rev Immunol. 14:141–153. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V, Bejnood A, Dionne D, Ge WH, Xu KH, et al: Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 184:4734–4752.e20. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Frede A, Czarnewski P, Monasterio G, Tripathi KP, Bejarano DA, Ramirez Flores RO, Sorini C, Larsson L, Luo X, Geerlings L, et al: B cell expansion hinders the stroma-epithelium regenerative cross talk during mucosal healing. Immunity. 55:2336–2351.e12. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang X, Hu C, Huang C, Wei Y, Li X, Hu M, Li H, Wu J, Czajkowsky DM, Guo Y and Shao Z: Robust Acquisition of spatial transcriptional programs in tissues with immunofluorescence-guided laser capture microdissection. Front Cell Dev Biol. 10:8531882022. View Article : Google Scholar : PubMed/NCBI | |
|
Ghaddar B and De S: Reconstructing physical cell interaction networks from single-cell data using Neighbor-seq. Nucleic Acids Res. 50:e822022. View Article : Google Scholar : PubMed/NCBI | |
|
Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, Bai C, Mo YY and Zhang Z: Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. 15:21667002023. View Article : Google Scholar : PubMed/NCBI | |
|
Sun C, Wang A, Zhou Y, Chen P, Wang X, Huang J, Gao J, Wang X, Shu L, Lu J, et al: Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer. Nat Commun. 14:26922023. View Article : Google Scholar : PubMed/NCBI | |
|
Tun HM, Peng Y, Massimino L, Sin ZY, Parigi TL, Facoetti A, Rahman S, Danese S and Ungaro F: Gut virome in inflammatory bowel disease and beyond. Gut. 73:350–360. 2024. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao E, Stone MR, Ren X, Guenthoer J, Smythe KS, Pulliam T, Williams SR, Uytingco CR, Taylor SEB, Nghiem P, et al: Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 39:1375–1384. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Saviano A, Henderson NC and Baumert TF: Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J Hepatol. 73:1219–1230. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S and Klappenbach JA: Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 35:936–939. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Thornton CA, Mulqueen RM, Torkenczy KA, Nishida A, Lowenstein EG, Fields AJ, Steemers FJ, Zhang W, McConnell HL, Woltjer RL, et al: Spatially mapped single-cell chromatin accessibility. Nat Commun. 12:12742021. View Article : Google Scholar : PubMed/NCBI | |
|
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert P: Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 14:865–868. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Vickovic S, Lötstedt B, Klughammer J, Mages S, Segerstolpe Å, Rozenblatt-Rosen O and Regev A: SM-omics is an automated platform for high-throughput spatial multi-omics. Nat Commun. 13:7952022. View Article : Google Scholar : PubMed/NCBI | |
|
Park HE, Jo SH, Lee RH, Macks CP, Ku T, Park J, Lee CW, Hur JK and Sohn CH: Spatial transcriptomics: Technical aspects of recent developments and their applications in neuroscience and cancer research. Adv Sci (Weinh). 10:e22069392023. View Article : Google Scholar : PubMed/NCBI | |
|
Rao A, Barkley D, França GS and Yanai I: Exploring tissue architecture using spatial transcriptomics. Nature. 596:211–220. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang L, Chen D, Song D, Liu X, Zhang Y, Xu X and Wang X: Clinical and translational values of spatial transcriptomics. Signal Transduct Target Ther. 7:1112022. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Stanojevic S and Garmire LX: Emerging artificial intelligence applications in spatial transcriptomics analysis. Comput Struct Biotechnol J. 20:2895–2908. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Hao M, Hua K and Zhang X: SOMDE: A scalable method for identifying spatially variable genes with self-organizing map. Bioinformatics. 37:4392–4398. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Dries R, Zhu Q, Dong R, Eng CL, Li H, Liu K, Fu Y, Zhao T, Sarkar A, Bao F, et al: Giotto: A toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22:782021. View Article : Google Scholar : PubMed/NCBI |









