Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer
- Authors:
- Published online on: May 15, 2014 https://doi.org/10.3892/or.2014.3186
- Pages: 395-402
Abstract
Introduction
Colon cancer is one of the most frequently diagnosed cancer and the third most fatal malignancy worldwide (1). Growing urbanization and industrialization have led to the steady rise in colon cancer incidence in China during the last 20 years (2). In certain high incidence areas, colon cancer has become the second leading cause of cancer-related mortality. Surgical resection is widely considered as the most useful therapy for colon cancer. Tumor metastasis is the major cause for the recurrence of colon cancer and leads to the failure of colon cancer treatment following radical operation. At present, clinicopathologic staging is the main risk assessment for colon cancer metastasis. Clinical outcomes, however, are quite variable, even among patients at the same stage. Therefore, research focusing on the molecular basis of colon tumor metastasis with the aim of implementing individualized therapeutic regimens for patients with colon cancer is urgently required.
The complicated process of tumor metastasis involves a number of molecular biological changes, including genetic and epigenetic mutations (3). Recently, the contribution of long non-coding RNAs in cancer progression has attracted increased attention. Long non-coding RNAs (lncRNAs) (typically >200 nt), a type of regulator non-coding RNAs, are increasingly reported to play key roles in numerous biological processes such as development, differentiation, diseases via integrity of nuclear structure, chromatin remodeling and post-transcription regulation (4). Gupta et al (5) first discovered that HOTAIR (Hox transcript antisense intergenic RNA) is one of the cancer metastasis-associated lncRNAs, and is a powerful biomarker of breast cancer metastasis and poor prognosis. Many other groups successively reported that HOTAIR overexpression is linked to the malignant features of many other cancer types (6–13). Moreover, Rinn et al (14) and Tsai et al (15) suggested that HOTAIR lncRNA serves as a scaffold for at least two histone modification complexes in order to induce transcriptional silencing of hundreds of genes including tumor-suppressor genes. Nevertheless, the mechanism of how HOTAIR is involved in colon cancer progression remains unknown.
In the present study, we evaluated the expression of HOTAIR in colon cancer tissue and paired normal mucosa and metastasized lymph node, and then determined the correlation between HOTAIR expression and clinicopathologic characteristics. Using RNA interference experiments, we ascertained whether HOTAIR has effects on colon cancer cell biological properties. Furthermore, we revealed how HOTAIR initiates epithelial-mesenchymal transition of colon cancer, the main mechanistic step resulting in metastasis.
Materials and methods
Tissue samples and cell lines
Samples from 120 patients undergoing colectomy between October 2007 and December 2010 by the same surgical team at the Shanghai Jiao Tong University Affiliated First People’s Hospital were collected and archived for the study. Fresh colon cancer samples and matched adjacent non-tumor tissues were obtained from 52 males and 68 females with ages ranging from 35 to 88 years. The samples were stored at −80°C immediately following removal. None of the patients had received any preoperative therapy, and patients with stage II, III and IV disease underwent standard adjuvant chemotherapy according to The National Comprehensive Cancer Network Practice Guidelines for Colon Cancer (1). Follow-up was carried out completely (median follow-up time, 55.5 months; range, 10–72 months). The study was approved by the Ethics Committee of Shanghai Jiaotong University Affiliated First People’s Hospital. Informed written consent was obtained from each patient enrolled in our study according to the Declaration of Helsinki. Colon cancer cell lines were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere containing 5% CO2 at 37°C.
RNA isolation and quantitative RT-PCR for assessment of mRNA expression
Total RNAs from all samples were isolated with AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany). After the RNA integrity and purity were checked, the first single-stranded cDNAs were synthesized from 3 μg of RNA, according to the manufacturer’s instructions (Fermentas, Shenzhen, China). One microliter of the cDNAs from each sample was used as a template for qRT-PCR analysis with the ABsolute qPCR SYBR-Green Mix (Fermentas), employing the ABI Prism 7900 system (Applied Biosystems Inc., Foster City, CA, USA).
The following primers were used for qRT-PCR: HOTAIR sense 5′-GGTAGAAAAAGCAACCACGAAGC-3′ and antisense 5′-ACATAAACCTCTGTCTGTGAGTGCC-3′; GAPDH sense 5′-GGAGCGAGATCCCTCCAAAAT-3′ and antisense 5′-GGCTGTTGTCATACTTCTCATGG-3′.
The expression level of HOTAIR was normalized to the transcription level of GAPDH. The quantitative PCR reaction for each sample was repeated in triplicate. The relative HOTAIR expression was calculated using the 2−ΔΔCt comparative method.
RNA interference
Two colon cancer cell lines were transfected with 50 nM positive siRNAs (si-HOTAIR 5′-GAACGGGAG UACAGAGAGAUU-3′) or negative control siRNA (Mock 5′-CUACAACAGCCACAACGUCdTdT-3′) employing Lipofectamine RNAiMAX (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions. Total RNA isolation was performed 72 h later for real-time PCR analysis, while other functional assays were carried out 48 h post-transfection. All RNA interference assays were carried out in triplicates independently.
Cell proliferation assay
For cell proliferation assay, cells were seeded in 96-well plates (2×103 cells/well) in triplicate. At the appropriate time (24, 48, 72, 96 and 120 h), the cells were incubated with MTT (5 mg/ml; Sigma, St. Louis, MO, USA) for 4 h at 37°C. MTT solution was then carefully removed and replaced with dimethyl sulfoxide (DMSO; Sigma). Absorbance was measured at a wavelength of 570 nm.
Cell migration and invasion assays
Cell migration and invasion assays were carried out using Transwell chambers (micropore size, 8 μm, 24-well; BD Biosciences, Franklin Lakes, NJ, USA) without Matrigel (for migration assay) or with Matrigel (for invasion assay). Both were performed according to the manufacturer’s protocol. Briefly, the treated cells were plated in the upper chamber at a concentration of 5×104 in 500 μl FBS-free media. The bottom chambers maintained suitable media with 10% FBS for 48 h. Then, the bottom of the chamber insert was fixed and stained with Giemsa solution. The stained cells were counted under a microscope at ×200 magnification; meanwhile, images were captured and saved. The experiments were carried out in triplicate.
Western blot analysis of protein expression
The total protein was extracted from cells using Thermo Scientific Pierce RIPA buffer combined with Thermo Scientific Halt proteinase inhibitor cocktail and then quantified using Thermo Scientific BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). The equivalent amounts of protein (20 μg) for each sample were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Membranes were blocked using 5% non-fat milk solution for 1 h, and then probed with primary polyclonal antibodies at 4°C overnight, followed by secondary antibodies (1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. β-actin was used as a loading control. After washing with TBST (Tris-buffered saline with 0.01% Tween-20), the proteins were detected by enhanced electrochemiluminescence (Millipore) and radiography. The primary antibodies used above were anti-E-cadherin (1:1,000; Abcam, Cambridge, UK) and anti-Vimentin (1:1,000; Abcam), anti-MMP9 (1:500; Cell Signaling Technology, Boston, MA, USA) and anti-β-actin antibody (1:1,000; Cell Signaling Technology).
Statistical analysis
All statistical analyses were set with a significance level of P≤0.05 and carried out using SPSS 19.0 statistical software (SPSS Inc., Chicago, IL, USA). The differences between 2 groups were analyzed with the Student’s t-test and χ2 test, as appropriate. The relationship between HOTAIR expression and clinicopathological characteristics was estimated with the Mann-Whitney U test and the Kruskal-Wallis test appropriately. Survival curves were calculated by Kaplan-Meier method, with the log-rank test employed for the comparison of differences. Variables estimated with significant means by univariate analysis were subjected to the multivariate Cox proportional hazards regression model.
Results
Overexpression of HOTAIR in colon cancer tissues
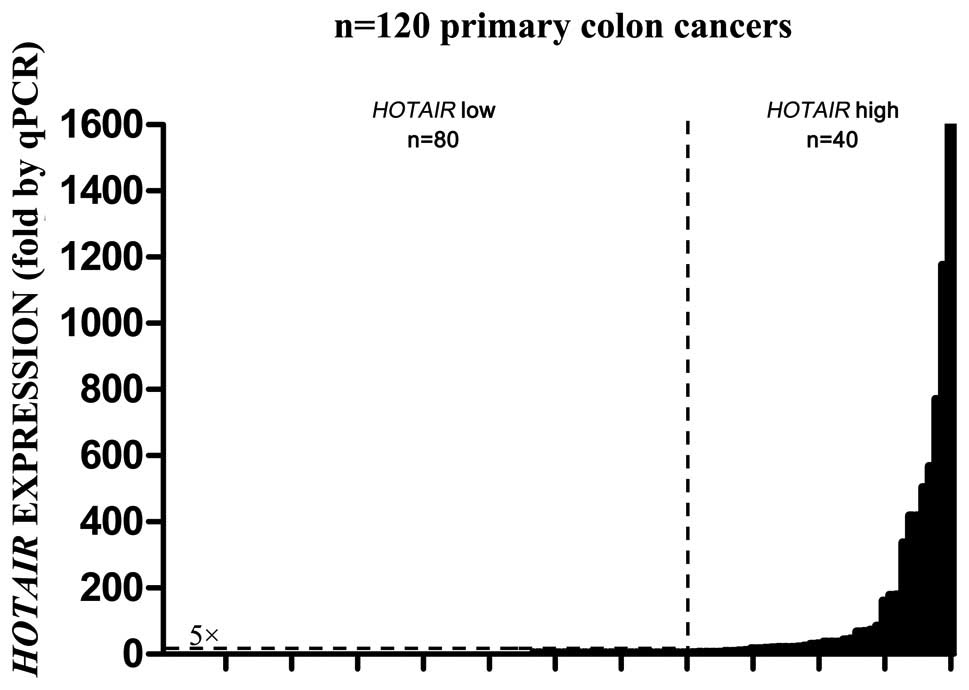
qPCR was applied to examine the expression level of HOTAIR in 120 pairs of cancer and matched normal tissues, and 32 metastasized lymph node tissues. As shown in Fig. 1, HOTAIR was overexpressed from 2- to nearly 1,600-fold in the cancer tissues. According to a HOTAIR expression level of >5-fold compared to normal tissues, 120 patients were divided into a high expression group (n=40) and a low expression group (n=80). In addition, 27 of 32 (84.4%) lymph node metastasis tissues showed high expression of HOTAIR, with significant means between primary cancer tissues and metastatic lymph node tissues (Table I).
Correlation between HOTAIR expression and clinicopathological characteristics
The correlation between HOTAIR expression and clinicopathological characteristics is summarized in Tables II and III. Increased HOTAIR expression was significantly associated with AJCC stage (P<0.001), lymph node metastasis (pN stage, P=0.002), organ metastasis (pM stage, P<0.001), vascular invasion (P=0.019) and histologic differentiation (P<0.001). No significant correlations were found between HOTAIR expression and age, gender or tumor location. The Kaplan-Meier curves (Fig. 2) revealed that patients in the high expression group had a significant poorer clinical prognosis than those in the low expression group, both for metastasis-free survival and overall survival (log-rank test P<0.001). With regard to Cox univariate proportional hazards regression model, the level of HOTAIR expression, lymph node metastasis, organ metastasis, AJCC stage, degree of differentiation and vascular invasion were prognostic factors. Additionally, multivariate analysis, including the variables above with P<0.05, indicated that HOTAIR expression was an independent clinical risk indicator for both metastasis-free survival (HR, 3.878; 95% CI, 1.369–10.981; P=0.011) and overall survival (HR, 3.915; 95% CI, 1.226–12.499; P=0.021).
Table IICorrelation between HOTAIR expression and clinicopathological characteristics in the colon cancer cases. |
Table IIIAssociation between clinical characteristics and metastasis-free and overall survival by Cox regression model analysis. |
HOTAIR promotes migration and invasion of colon cancer cells
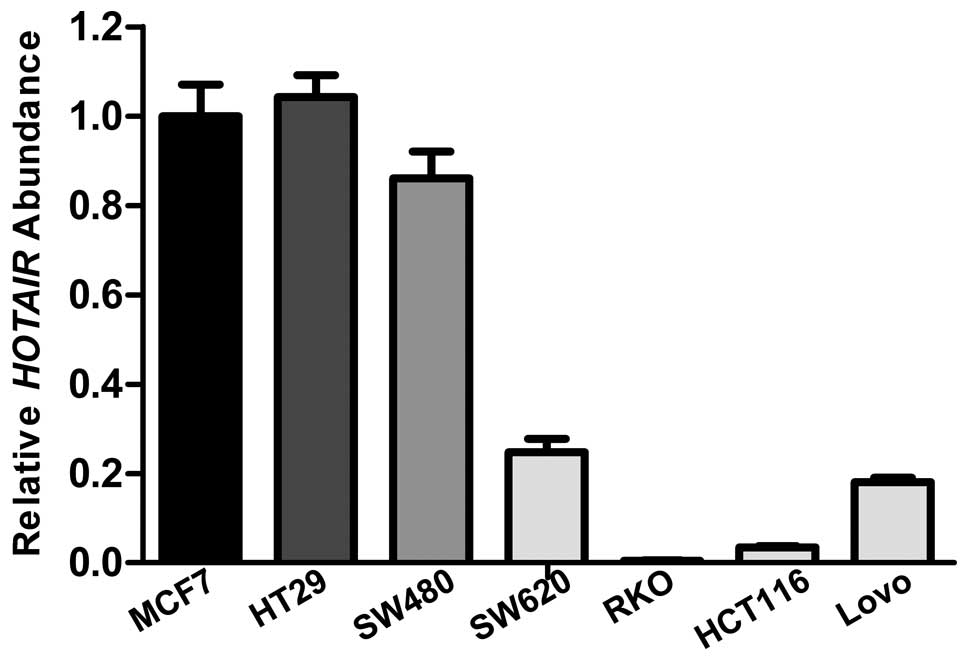
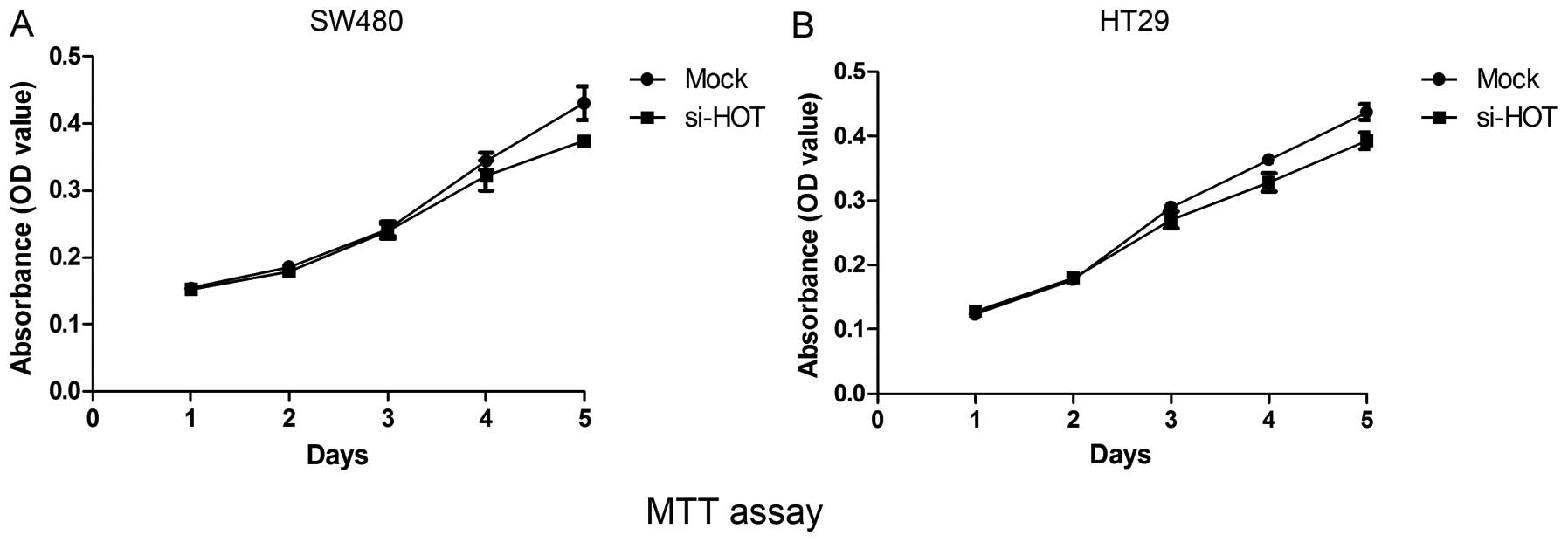
As reported by other research groups, HOTAIR may play a pro-oncogenic role in certain types of cancers such as breast (5), hepatocellular (6), gastrointestinal (8), pancreatic (9) and lung cancer (11). To examine the effect of HOTAIR on colon cancer, a series of functional experiments concerning cell proliferation and cell metastasis was performed. We quantified the expression levels of HOTAIR in the colon cancer cell lines by qRT-PCR, and compared the levels to the level in MCF-7, a breast cancer cell line that overexpresses endogenous HOTAIR (Fig. 3). HOTAIR expression levels in colon cancer cell lines SW480 and HT29 were higher than that in the other 4 cell lines. HOTAIR knockdown was carried out by using small interfering RNA transfection in the cell lines. The efficiency of siRNA-mediated knockdown in the SW480 or HT29 cells was >60% (data not shown). As illustrated by Fig. 4, results from the MTT assay showed that the proliferation rate of cells depleted of HOTAIR was slightly slower than the rate of cells treated with mock siRNA throughout 5 continuous days.
We next ascertained whether manipulation of the HOTAIR level affects cell metastasis in vitro. Migration and invasiveness of tumor cells are key aspects of metastasis. In the Transwell migration assay, fewer SW480 and HT29 colon cancer cells treated by si-HOTAIR migrated to the 8-μm pores of the Transwell chambers when compared to the cells treated with Mock (Fig. 5A and B). Furthermore, similar results were obtained in the invasion assay using a Matrigel environment study (Fig. 5C and D). All of these studies were repeated three times and the differences were statistically significant (P<0.05). Based on these outcomes, it was suggested that HOTAIR is involved in the progression of colon cancer mainly by promoting cancer cell migration and invasiveness.
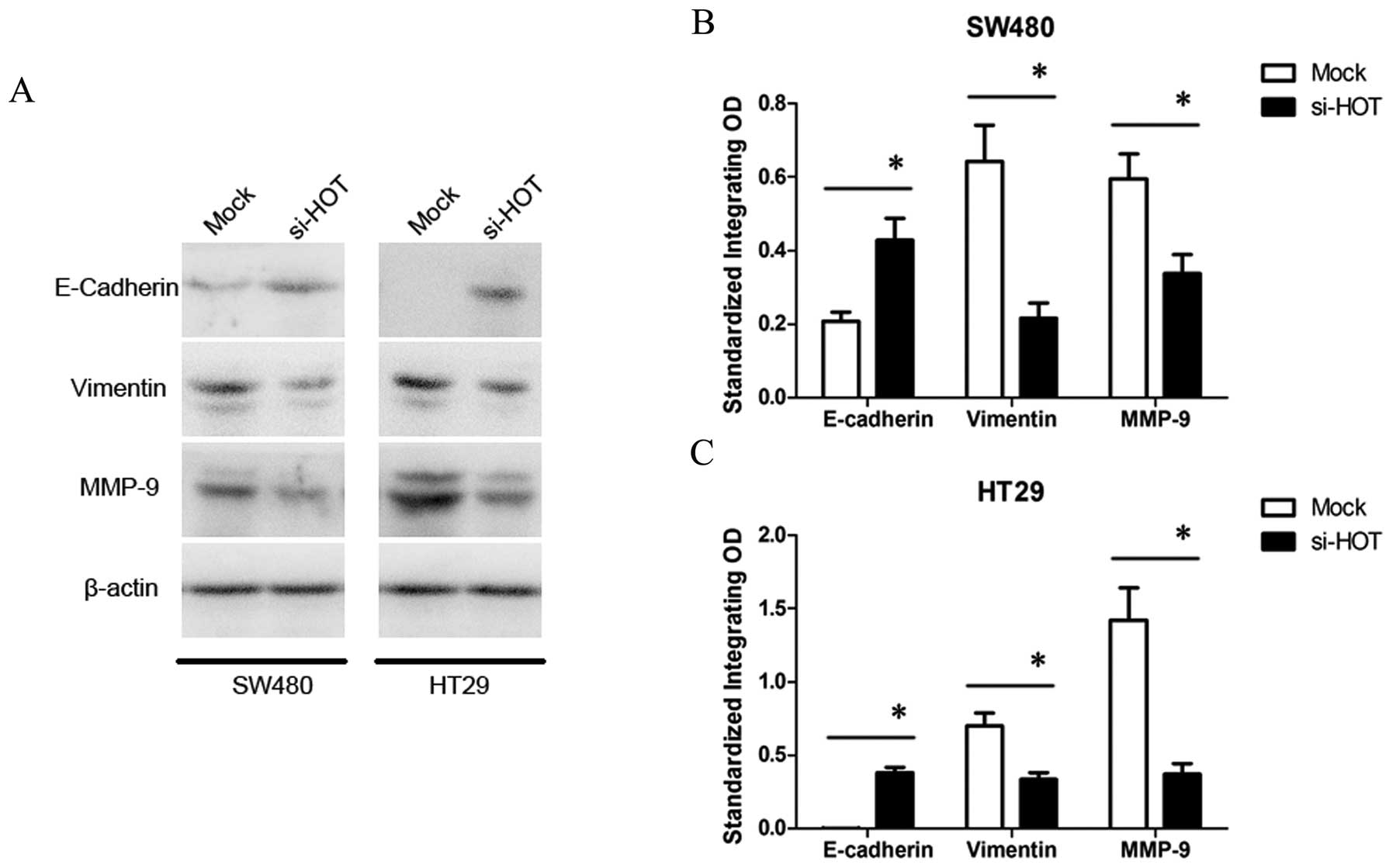
HOTAIR regulates expression of E-cadherin, vimentin and MMP9
Epithelial-mesenchymal transition (EMT) with alterations in gene expression profiles of cells initiates the invasion-metastasis cascade. Therefore, we ascertained whether HOTAIR promotes EMT to regulate tumor progression. Several key genes involved in EMT of colon cancer were selected for further western blot analysis (Fig. 6). When SW480 and HT29 cells were treated by HOTAIR knockdown, expression of E-cadherin, a hallmark of epithelial cell was increased, while expression of vimentin, a mesenchymal cell marker, was decreased. Additionally, in the si-HOTAIR-treated cells, expression of matrix metalloproteinase 9 (MMP9), which functions as an important proteinase to degrade the extracellular matrix for facilitating cell motility, was inhibited as well. These data indicate that the regulation of EMT is partly involved in the mechanism of HOTAIR-induced metastasis.
Discussion
HOTAIR lncRNA, with a 2158-nucleotide transcript, is transcribed from the HOXC locus in an antisense manner, which is also the reason why it is termed HOXC antisense intergenic RNA (14). HOTAIR has very high nucleotide conservation in vertebrates and an anatomic conservative expression pattern from embryo to adulthood. Moreover, the lncRNA is selectively required to physically interact with polycomb repressive complex 2 (PRC2) to regulate the chromatin methylation state. PRC2, comprising H3K27 histone methyl transferase EZH2, SUZ2 and EED, is involved in stem cell pluripotency and cancer progression (16–18). In 2010, HOTAIR was reported as a powerful predictor of breast cancer metastasis and poor prognosis. Gupta and his coworkers (5)found a positive association among high expression pattern of HOTAIR, breast cancer malignant biologic characteristics and reduced survival. The present study showed that the level of HOTAIR lncRNA expression in tumor tissues was distinctly higher than the level in paired normal mucosa. Additionally, HOTAIR was more highly expressed in invaded lymph nodes than in primary cancer tissues. Increased HOTAIR expression was significantly correlated with the depth of tumor invasion, lymph node metastasis, organ metastasis, histological differentiation, vascular invasion and advanced tumor stage. Patients with high HOTAIR expression had higher recurrence rates and poorer metastasis-free and overall survival rates than patients with low HOTAIR expression. Multivariate analyses revealed that HOTAIR expression was an independent factor for colon cancer prognosis after surgery. These results indicated that HOTAIR also could be a meaningful predictor for colon cancer management.
The 5′ domain of HOTAIR binds to PRC2 and is required for histone H3 lysine 27 trimethylation in order to silence HOXD and other target genes (5,14). On the other hand, the 3′ domain of HOTAIR maintains LSD1-binding activity (15). LSD1, the first identified demethylase that specifically catalyzes the demethylation of histone H3 lysine 4, is believed to be linked to the transcriptional silencing of tumor-suppressor genes (19–22). Tsai et al (15) suggested that HOTAIR, as a modular scaffold, is required to target both PRC2 and LSD1 to many promoter elements of genes in order to coordinate chromatin modification for gene repression. Loss of HOTAIR notably weakens the activities of PRC2 and LSD1 in modifying the histone state. Therefore, it is reasonable to expect that HOTAIR plays a crucial role in cancer development other than a merely novel cancer biomarker. At present, the study results revealed that HOTAIR had a limited effect on cell proliferation. As HOTAIR knockdown using siRNA significantly reduced the ability of cancer cells to invade the Transwell membranes, this lncRNA may promote colon cancer cell migration and invasion, which is mainly consistent with previous studies in other cancer types.
We further addressed how HOTAIR-dependent gene regulation in colon tumors is involved in the enhancement of an aggressive biological behavior. The early step leading to metastasis is migration and invasion of cancer cells from original tissues into the surrounding stroma. In order to acquire such abilities, carcinoma cells must inherit a drastic phenotypic alteration - the epithelial-mesenchymal transition (EMT). EMT is a biological process that allows polarized epithelial cells to experience multiple biochemical changes that enable them to assume a mesenchymal cell phenotype (23). Upon the phenotype change, cancer cells lose intercellular junction, breach the basement membrane and acquire enhanced motility. The complexity of EMT means that only small numbers of centrally pleiotropic regulators orchestrate the complement, such as Twist, Slug and Snail and the miR200 family. The present study demonstrated that depletion of HOTAIR increased expression of E-cadherin while concomitantly decreasing expression of vimentin and MMP9. Together with previous studies using global gene expression analysis, the emerging evidence indicates that increased expression of HOTAIR in cancer cells appears to reprogram the chromatin profile of epithelial cells to that of mesenchymal cells, cooperating with PRC2 and LSD1 at least. Hence, HOTAIR may be another pleiotropic modulator participating in EMT.
Collectively, our data indicate that HOTAIR is a valuable factor for colon cancer prognosis. Moreover, HOTAIR can promote colon cancer cell migration and invasiveness and may participate in epithelial-mesenchymal transition. Further studies are warranted to advance our understanding of the involvement of HOTAIR in cancer development, since this lncRNA is a potential target for cancer prevention and treatment.
Acknowledgements
The present study was supported by funds from the National Natural Science Foundation of China (81172328), the Important International Cooperation grants from the National Natural Science Foundation of China (81220108021), the Medical Guidance Project of Shanghai Science and Technology Commission (124119a1700), the Science and Technology Innovation Plan of Shanghai Science and Technology Commission (11431921000), the Frontier Technology Union Research Project of Shanghai Municipal Hospitals (SHDC12012105), and the Medical Climbing Project from Songjiang Health Bureau of Shanghai (2011PD03).
References
|
NCCN Clinical Practice Guidelines in Oncology for Colon Cancer (Version 3). 2013, [EB/OL] Available at www.nccn.org/professionals/physician_gls/f_guidelines.asp. | |
|
Li M and Gu J: Changing patterns of colorectal cancer in China over a period of 20 years. World J Gastroenterol. 11:4685–4688. 2005.PubMed/NCBI | |
|
Fearon ER: Molecular genetics of colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Ulitsky I and Bartel DP: lincRNAs: genomics, evolution, and mechanisms. Cell. 154:26–46. 2013. View Article : Google Scholar | |
|
Gupta RA, Shah N, Wang KC, et al: Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. 2010.PubMed/NCBI | |
|
Yang Z, Zhou L, Wu LM, et al: Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 18:1243–1250. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Lu L, Zhu G, Zhang C, et al: Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 136:875–883. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Niinuma T, Suzuki H, Nojima M, et al: Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72:1126–1136. 2012.PubMed/NCBI | |
|
Kim K, Jutooru I, Chadalapaka G, et al: HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Li D, Feng J, Wu T, et al: Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar | |
|
Nakagawa T, Endo H, Yokoyama M, et al: Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013. View Article : Google Scholar | |
|
Zhuang Y, Wang X, Nguyen HT, et al: Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 6:352013. View Article : Google Scholar : PubMed/NCBI | |
|
Kogo R, Shimamura T, Mimori K, et al: Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71:6320–6326. 2011.PubMed/NCBI | |
|
Rinn JL, Kertesz M, Wang JK, et al: Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Tsai MC, Manor O, Wan Y, et al: Long noncoding RNA as modular scaffold of histone modification complexes. Science. 329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Sparmann A and van Lohuizen M: Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 6:846–856. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Richly H, Aloia L and Di Croce L: Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2:e2042011. View Article : Google Scholar : PubMed/NCBI | |
|
Crea F, Fornaro L, Bocci G, et al: EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 31:753–761. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Ding J, Zhang ZM, Xia Y, et al: LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 109:994–1003. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Huang Z, Li S, Song W, et al: Lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/β-catenin pathway by down-regulating Dickkopf-1 (DKK1). PLoS One. 8:e700772013.PubMed/NCBI | |
|
Tang M, Shen H, Jin Y, et al: The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. J Biol Chem. 288:27680–27691. 2013. View Article : Google Scholar | |
|
Abdel-Wahab O and Dey A: The ASXL-BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia. 27:10–15. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Kalluri R and Weinberg RA: The basics of epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428. 2009. View Article : Google Scholar : PubMed/NCBI |