The emerging landscape of circular RNA ciRS-7 in cancer (Review)
- Authors:
- Published online on: April 8, 2015 https://doi.org/10.3892/or.2015.3904
- Pages: 2669-2674
Abstract
Introduction
Cancer is a leading cause of mortality that contributes the highest number of deaths from diseases. There were 14.1 million new cases and 8.2 million deaths worldwide in 2012 based on the GLOBOCAN series of the International Agency for Research on Cancer (1). At the time of diagnosis, most of the patients have developed invasive cancer. At the same time, a large number of trials have been performed to find an effective treatment strategy, but the overwhelming number of various cancers still carry high risks of morbidity and mortality (2,3). Therefore, there is a great need and urgency in the search for early diagnosis biomarkers and novel treatment measures to improve the survival and the cure rate of cancer.
Genome-wide association analyses have shown that in humans, over half of all miRNAs are located at chromosomal fragile sites, which are associated closely with malignant tumors (4,5). This suggests that miRNAs play an important role in the development of malignant tumors. For example, the expression of microRNA-7 (miR-7) is downregulated in tumor tissues or cancerous cells found in breast cancer (6), tongue cancer (7), gastric carcinoma (8), lung neoplasm (9), hepatocellular carcinoma (HCC) (10), schwannoma (11) and cervical cancer (12). It also plays a significant role in invasion and metastasis and other tumor biological processes. In March 2013, two publications reported that circular RNAs (circRNAs) function as miRNA ‘sponges’ that act to naturally sequester and competitively suppress the activity of miRNAs (13,14). These findings suggest that circRNAs may play a role in cancer.
CircRNAs are a large class of non-coding RNAs that exist ubiquitously in the cytoplasm of eukaryotic cells (15). At present, they are the subject of intense research in the field of RNAs. Recently, some circRNAs have been identified, including circRNA sponge for miR-7, known as ciRS-7. ciRS-7 is also named cerebellar degeneration-related protein 1 antisense RNA (CDR1as) or CDR1NAT (13,16). It is ~1,500 nucleotides in length and is found predominantly in human and mouse brain (16), where it naturally occurs as an antisense RNA strand and takes on the structure of a closed loop. As a super sponge or competing endogenous RNA (ceRNA) of miR-7, ciRS-7 competitively sequesters and quenches the activity of miR-7 (13,14,17). This has significant implications for the biology of cancer, as miR-7 has tremendous effects on the development of a variety of cancer, such as breast cancer, hepatocellular carcinoma, and cervical cancer (12). To address the question whether ciRS-7 is also implicated in cancer, we will review the origin and functions of circRNAs and in particular, ciRS-7, and discuss the connection between ciRS-7 and its molecular targets that are involved in cancer. It is our hope that further research in this area may provide a new direction toward the early diagnosis and targeted therapy of cancer.
2. Origin of circRNAs
CircRNAs are ubiquitous in the cytoplasm of eukaryotic cells, and have returned to the forefront of research with the discovery of their abundance thanks to technological breakthroughs in high-throughput deep sequencing. In 1979, scientists observed for the first time the presence of small RNA structural variants that were circular in shape in the cytoplasm of eukaryotic cells by electron microscope (18). These were mistaken as rare existing transcriptional ‘noise’ (19). However, with the emergence of high-throughput sequencing and RNA-seq, several studies have confirmed the widespread existence of circRNAs in eukaryotic organisms and their role in carrying out important regulatory functions. The question is then, how are circRNAs generated?
CircRNAs may be generated from exons (exonic circRNA) or introns (intronic circRNA). Jeck et al put forth two models for the origination of circRNAs, namely lariat-driven circularization and intron-pairing-driven circularization (20,21). The main difference is in the first step of circRNAs generation. The first model requires the covalent binding of the splice donor with the splice acceptor in the exon, while the second model involves complementary pairing within two introns to form a circular structure. The subsequent steps of circRNA formation are largely similar. Namely, the spliceosome excises residual introns and forms the circRNAs.
In addition, recent studies have shown that there is an alternative circularization pathway for the production of circRNAs. This alternative circularization of RNAs relies on the human genome containing a large number of complementary sequences (such as Alu sequences) in the intronic regions. The selective pairing and dynamic regulation of these complementary sequences allows the same gene to produce multiple circRNAs (22,23). However, not all of the complementary sequences can produce circularization. The production of circRNAs is facilitated by short intronic repeat sequences, while it is suppressed by increasing hairpin stability between sequences (24). Endogenous conditions of circRNAs formation are very complicated. For example, the number and distance of complementary sequences across the circularization exons can influence matching and competition between complementary sequences. Alternative circularization is closely related to normal physiological function, while aberrant alternative circularization leads to a variety of important human diseases (22), such as cancer.
3. Functions of ciRS-7
miRNA sponges
There are thousands of circRNAs expressed at different positions in the genome of various species, from archaea to humans (25–28). Nearly 2,000 types of circRNAs have been found in humans, 900 types have been identified in mice, and ~700 types exist in nematodes based on sequencing data (14,29). Among these, ciRS-7 expression shows complex tissue specificity, cell type specificity, and developmental stage specificity. It has been reported that (14): i) ciRS-7 binds densely to the miR-7 effector; ii) ciRS-7 contains 74 binding sites for miR-7, 63 of which exist in at least one other organism; iii) ciRS-7 and miR-7 possess the same specific expression region in the embryonic brain tissue of mice; iv) ciRS-7 is circularized and is not detectable as a linear RNAs; v) ciRS-7 injection shows similar phenotypes as miR-7 knock-out in zebrafish brains. The above-described data show that ciRS-7 acts as a ceRNA of miR-7 and functions as a natural inhibitor of miR-7 and competes miR-7 with other RNAs, such as mRNAs, and lncRNAs (Fig. 1A).
Transcriptional and posttranscriptional regulations
Artificial circRNAs contain an internal ribosome entry site that can be translated in the cellular extract (30), but endogenous circRNAs have not yet been found to be associated with ribosomes. Hence, linear RNAs and circRNAs must be closely regulated, as their presence can radically change the functional output of a gene (20,27). Owing to its ability to sequester miR-7, ciRS-7 plays an important role in fine-tuning of the expression of target genes by binding in trans to the 3′UTRs of a target mRNA (Fig. 1B). The combination of miR-7 with ciRS-7 may even silence the trans-acting activity. Stable expression of ciRS-7 in HeLa cells results in significantly lower expression of α-synuclein (SNCA), epidermal growth factor receptor (EGFR), and IRS2 mRNA (13). This result suggests that ciRS-7 presumably regulates the expression of SNCA, EGFR, and IRS2 by a posttranscriptional mechanism in HeLa cells. ciRS-7 may be associated with the assembly of large RNAs or protein complexes, perhaps similar to other molecules with low complexity (31). In addition, ciRS-7 acts as an ‘mRNA trap’ by forming closed translation initiation sites (21), and leaves the non-coding linear transcript, thereby reducing the expression levels of protein.
Interaction with binding protein
CiRS-7 may act as ‘scaffolding’ for RNA binding protein (RBP) to combine multiple protein subunits (21,32) and to facilitate the interaction by potentially increasing the stability of the circRNAs transcripts. The capturing mechanism of miRNA sponges is equally important to RBP. Similarly, ciRS-7 can store, classify or find RBP. Besides, ciRS-7 also interacts widely with the argonaute (AGO) protein (14) (Fig. 1C).
To summarize, ciRS-7 can regulate the activities of miRNAs, mRNAs, and RBP by competing miRNAs with other RNAs molecules, by binding in trans with the 3′UTRs of target gene mRNAs, and by interacting with proteins, respectively (Fig. 1). These are the mechanisms by which ciRS-7 exerts its specific biological effects.
4. ciRS-7 and cancer
miR-7 and cancer
miRNAs are a group of single-stranded small non-coding RNAs consisting of 19–24 nucleotides, which regulate gene expression by interfering with the posttranscriptional process (33,34). In the past 10 years, growing evidence suggests that abnormal expression of miRNAs is a sign of malignant tumors (35,36), as miRNAs can affect cell proliferation, migration, apoptosis, and much other cancer-related biological process. miRNAs involved in cancer can be divided into oncomiRs and tumor suppressor miRNAs. As a tumor suppressor miRNA, the absence/low expression or dysfunction of miR-7 participates in the initiation and evolution of various malignancies in humans, including breast cancer, hepatocellular carcinoma, and cervical cancer. Liu et al found that (12) miR-7 can inhibit the growth of cancer cells and promote apoptosis, and can, therefore potentially be a treatment target in cancer.
miRNAs can prevent or promote the translation and degradation of target gene by binding to the 3′UTR of target gene mRNAs, thereby regulating the expression of target genes (37). However, miRNA-mediated upregulation of target genes is usually specific and depends on distinct target mRNAs (38). It has been proven that target genes of miR-7 mainly including EGFR, X-linked inhibitor of apoptosis protein (XIAP), Kruppel-like factor 4 (KLF4), SNCA, IRS2, IRS1, 4EBP1, ACK1, AKT, FAK, HNF4, IGF1R, mTOR, NOTCH1, RAF1, p70S6K, PA28c, PAK1, and PIK3CD, all of which play indispensable roles as oncogenes or tumor suppressor genes in a variety of cancers.
In cervical cancer, as an example, EGFR is constitutively highly expressed in pancreatic cancer, oral cancer, cervical cancer and other cancers, and participates in the pathogenesis and progression of these cancers (39–41). The variation in EGFR regulates the risk of developing in situ and invasive cervical cancer (42); while inhibition of EGFR results in antitumor effects and can strengthen the curative effect of chemoradiotherapy based on cisplatin (43). As an oncogene, XIAP expression is increased in cervical cancer cells, and regulated positively by TGF-β in a TGF-β/Smad signaling pathway-dependent manner (44). It is associated closely with survival of cervical cancer patients and promotes oncogenesis. Also, it regulates the sensitivity of human cervical cancer cells to cisplatin, doxorubicin, and taxol (45). As a transcription factor of zinc finger-like structure, KLF4 can inhibit the growth of cancer cells and tumor formation (46,47). Thus, it functions as a tumor suppressor in cervical cancer. The above research suggests that both oncogenes (such as EGFR and XIAP) and tumor suppressor genes (such as KLF4) can be treated as target molecules of miRNAs, and play a vital role in the initiation, development, and prognosis of cancer (such as cervical cancer).
ciRS-7 and cancer
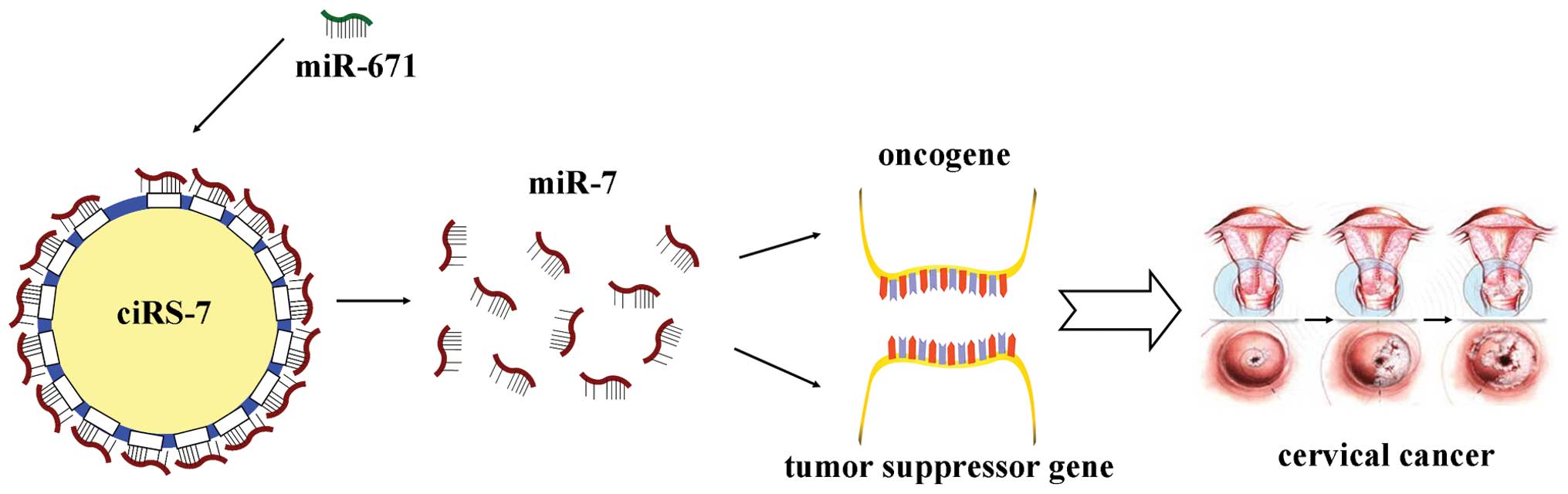
It has been reported that a circular isoform of the non-coding RNA ANRIL is associated with the risk for the development of atherosclerosis (15,48). This finding indicates that circRNAs have biochemical and phenotypic consequences. Recently, the expression of ciRS-7 was found to be decreased in the hippocampus of Alzheimer’s patients, suggesting that there is a deficiency in the ‘sponging’ effects of ciRS-7 when the level of miR-7 is increasing and the expression of miR-7-sensitive mRNA targets is decreasing (49). It has been implied that ciRS-7 likely serves as a crucial factor that is significantly involved in various diseases. In addition, a recent review reported that expression analyses of various tumor cell lines showing wide spread expression of ciRS-7 in neuroblastomas and frequent expression in astrocytoma, renal cell, and lung carcinomas (50). The stable expression of ciRS-7 in HeLa cells (13,15) indicates that ciRS-7 may be associated with cervical cancer. There is a large degree of subcellular co-localization between ciRS-7 and miR-7 in HeLa cells when analyzed by RNA-fluorescence in situ hybridization and IF-fluorescence in situ hybridization (13). Moreover, ciRS-7 can be used as an ideal ceRNA of miR-7, as it sequesters and potently quenches the normal activities of miR-7 (14). miR-7 is involved in multiple biological processes of various cancers; for example, it promotes the growth of cervical cancer cells. Furthermore, ciRS-7 is sensitive to miR-671. miR-671 can induce the degradation and endonucleolytic cleavage of ciRS-7, so ciRS-7 may be responsible for bringing miR-7 to a subcellular location where miR-671 promotes the release of miR-7 by ciRS-7 (14,51). Thus suggesting that miR-671 may inhibit the expression of miR-7. From the interaction of ciRS-7 with cancer associated miR-7, we surmise that they have a potential role in the regulation of cancer (such as cervical cancer).
In summary, ciRS-7 may act as a ceRNA of miR-7, competitively inhibiting the activity of miR-7, promotes the expression of oncogenes (such as EGFR and XIAP), while inhibiting tumor suppression genes (such as KLF4), therefore promoting the initiation and development of cancer (take cervical cancer for example) (Fig. 2).
5. Potential values of circRNAs in diagnosis and treatment of cancer
As diagnostic and prognostic biomarkers
According to the current studies, the main characteristics of circRNAs are as the following. i) Universality: a report in 2012 confirmed for the first time that circRNAs are the most common molecules after linear RNAs in human cells (15) and that they are far more ubiquitous than previously imagined. ii) Conservatism: the signal behind circularization seems to be evolutionarily conserved in different species (52); for example, some circRNAs are detected in both humans and mice. iii) Cytoplasmic location. iv) Definite specificity (53). v) Stability: the half-life of circRNAs in most species is longer than 48 h, while the average half-life of mRNAs is 10 h (21). This may be attributable to the resistance of circRNAs to ribonuclease (RNaseR). Nucleases usually recognize the end of linear RNA molecules, and the closed structure of circRNAs likely grants them their high tolerance to nucleases. vi) Highly abundant expression: the gene product level of some exonic circRNAs has been shown to be higher than that of linear RNAs as measured by the sequencing method and quantitative PCR (21,54). Sometimes, the abundance of circRNAs is over 10 times that of the corresponding linear mRNAs. This abundance may be attributed to the stability of circRNAs. Therefore, circRNAs possess distinct advantages and have the potential to be biomarkers of diagnosis and prognosis in cancer.
Contribution to targeted therapy
Circularization may be the future target of treatment, either to reduce the circularization of functional transcripts, or to sequester by an ‘mRNA trap’, exons contributing to dysfunctional transcripts (21). Compared to the miRNA sponges that only contain a string of single miRNA response elements (MREs), ciRS-7 contains plenty of MREs that bind miR-7 (51) (Fig. 1A). As such, it can instantaneously bind or release a large number of miR-7 molecules, thereby effectively regulating the network of diseases. In comparison with single miRNA-targeted technology, targeting inhibition of the expression of ciRS-7 can amplify the protective effect of the tumor suppressor miR-7. Compared with single RNAi technology targeting oncogenes, inhibiting the expression of ciRS-7 could affect the activities of multiple oncogenes. In short, ciRS-7 contains a large number of MREs, can regulate the expression of several targeted molecules, and it possesses more advantages in comparison with a single miRNA or single gene.
In conclusion, as a ceRNA of miR-7, ciRS-7 can naturally sequester and inhibit the activity of miR-7 and promote the expression of oncogenic EGFR and XIAP gene as well as suppress the expression of tumor-suppressed KLF4, thus promoting the initiation and development of cancer (cervical cancer was is shown as an example) (Fig. 2). However, the biological functions and its mechanism still have to be validated and further investigated. Although studies on circRNAs are just beginning, the significance in diseases/cancers has gradually grown. With unique characteristics as biomarkers owing to its stability and specificity, ciRS-7 can provide a new direction to improve the early screening and diagnosis in cancer. Also, ciRS-7 possesses unique characteristics as an ideal ceRNA, which could provide a novel avenue for the development of treatments against cancer.
Acknowledgments
This study was supported by National Key Basic Research Program (973 Program) funded project (2010CB833605), People’s Republic of China.
References
|
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015. View Article : Google Scholar | |
|
Dušek L, Mužík J, Malúšková D, Májek O, Pavlík T, Koptíková J, Gregor J, Brabec P and Abrahámová J: Epidemiology of screening-targeted cancers according to new data of the Czech National Cancer Registry. Klin Onkol. 27(Suppl 2): 19–39. 2014.In Czech. View Article : Google Scholar | |
|
Rosenberg AR, Kroon L, Chen L, Li CI and Jones B: Insurance status and risk of cancer mortality among adolescents and young adults. Cancer. Dec 9–2014.Epub ahead of print. PubMed/NCBI | |
|
Zhang X, Hu S, Zhang X, Wang L, Zhang X, Yan B, Zhao J, Yang A and Zhang R: MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 443:1078–1084. 2014. View Article : Google Scholar | |
|
Bartel DP: MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116:281–297. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, Wang A, Dai Y and Zhou X: MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 432:199–205. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Kong D, Piao YS, Yamashita S, Oshima H, Oguma K, Fushida S, Fujimura T, Minamoto T, Seno H, Yamada Y, et al: Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene. 31:3949–3960. 2012. View Article : Google Scholar | |
|
Li J, Zheng Y, Sun G and Xiong S: Restoration of miR-7 expression suppresses the growth of Lewis lung cancer cells by modulating epidermal growth factor receptor signaling. Oncol Rep. 32:2511–2516. 2014.PubMed/NCBI | |
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L: MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Saydam O, Senol O, Würdinger T, Mizrak A, Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky AM, Saydam N, et al: miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 71:852–861. 2011. View Article : Google Scholar : | |
|
Liu S, Zhang P, Chen Z, Liu M, Li X and Tang H: MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 587:2247–2253. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al: Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Salzman J, Gawad C, Wang PL, Lacayo N and Brown PO: Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 7:e307332012. View Article : Google Scholar : PubMed/NCBI | |
|
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statha AL, Clark SJ and Kjems J: miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Salmena L, Poliseno L, Tay Y, Kats L and Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Hsu MT and Coca-Prados M: Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI | |
|
Cocquerelle C, Mascrez B, Hétuin D and Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J. 7:155–160. 1993.PubMed/NCBI | |
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19:141–157. 2013. View Article : Google Scholar : | |
|
Jeck WR and Sharpless NE: Detecting and characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL and Yang L: Complementary sequence-mediated exon circularization. Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Vicens Q and Westhof E: Biogenesis of Circular RNAs. Cell. 159:13–14. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Liang D and Wilusz JE: Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Danan M, Schwartz S, Edelheit S and Sorek R: Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40:3131–3142. 2012. View Article : Google Scholar : | |
|
Wilusz JE and Sharp PA: Molecular biology. A circuitous route to noncoding RNA. Science. 340:440–441. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Guo JU, Agarwal V, Guo H and Bartel DP: Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI | |
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 9:e908592014. View Article : Google Scholar : PubMed/NCBI | |
|
Ghosal S, Das S, Sen R, Basak P and Chakrabarti J: Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 4:2832013. View Article : Google Scholar : PubMed/NCBI | |
|
Chen CY and Sarnow P: Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al: Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Ledford H: Circular RNAs throw genetics for a loop. Nature. 494:4152013. View Article : Google Scholar : PubMed/NCBI | |
|
Maqbool R, Ismail R and Hussain M: Mutations in microRNA genes and their binding sites are infrequently associated with human colorectal cancer in the Kashmiri population. MicroRNA. 2:219–224. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Macha MA, Seshacharyulu P, Krishn SR, Pai P, Rachagani S, Jain M and Batra SK: MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 20:5287–5297. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al: Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Di Leva G, Garofalo M and Croce CM: MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : | |
|
Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al: MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 51:846–856. 2010.PubMed/NCBI | |
|
Ma C, Qi Y, Shao L, Liu M, Li X and Tang H: Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life. 65:1026–1034. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Stock AM, Hahn SA, Troost G, Niggemann B, Zänker KS and Entschladen F: Induction of pancreatic cancer cell migration by an autocrine epidermal growth factor receptor activation. Exp Cell Res. 326:307–314. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Ribeiro FA, Noguti J, Oshima CT and Ribeiro DA: Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: A promising approach. Anticancer Res. 34:1547–1552. 2014.PubMed/NCBI | |
|
Tomao F, Di Tucci C, Imperiale L, Boccia SM, Marchetti C, Palaia I, Muzii L and Panici PB: Cervical cancer: Are there potential new targets? An update on preclinical and clinical results. Curr Drug Targets. 15:1107–1120. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Ma D, Hovey RL, Zhang Z, Fye S, Huettner PC, Borecki IB and Rader JS: Genetic variations in EGFR and ERBB4 increase susceptibility to cervical cancer. Gynecol Oncol. 131:445–450. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Nogueira-Rodrigues A, Moralez G, Grazziotin R, Carmo CC, Small IA, Alves FV, Mamede M, Erlich F, Viegas C, Triginelli SA, et al: Phase 2 trial of erlotinib combined with cisplatin and radiotherapy in patients with locally advanced cervical cancer. Cancer. 120:1187–1193. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Van Themsche C, Chaudhry P, Leblanc V, Parent S and Asselin E: XIAP gene expression and function is regulated by autocrine and paracrine TGF-beta signaling. Mol Cancer. 9:2162010. View Article : Google Scholar : PubMed/NCBI | |
|
Gagnon V, Van Themsche C, Turner S, Leblanc V and Asselin E: Akt and XIAP regulate the sensitivity of human uterine cancer cells to cisplatin, doxorubicin and taxol. Apoptosis. 13:259–271. 2008. View Article : Google Scholar | |
|
Yang WT and Zheng PS: Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis. PLoS One. 9:e888272014. View Article : Google Scholar : PubMed/NCBI | |
|
Yang WT and Zheng PS: Krüppel-like factor 4 functions as a tumor suppressor in cervical carcinoma. Cancer. 118:3691–3702. 2012. View Article : Google Scholar | |
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z and Sharpless NE: Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI | |
|
Lukiw WJ: Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 4:3072013. View Article : Google Scholar | |
|
Hansen TB, Kjems J and Damgaard CK: Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Kosik KS: Molecular biology: Circles reshape the RNA world. Nature. 495:322–324. 2013.PubMed/NCBI | |
|
AbouHaidar MG, Venkataraman S, Golshani A, Liu B and Ahmad T: Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 111:14542–14547. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Perkel JM: Assume nothing: The tale of circular RNA. Biotechniques. 55:55–57. 2013.PubMed/NCBI |