Oncolytic potential of an E4-deficient adenovirus that can recognize the stabilization of AU-rich element containing mRNA in cancer cells
- Authors:
- Published online on: November 12, 2018 https://doi.org/10.3892/or.2018.6865
- Pages: 954-960
Abstract
Introduction
For adenovirus replication, several early gene products, such as E1A, E1B and E4, are necessary to change the environment within host cells (1). The E4 region of adenoviruses encodes multiple proteins and these proteins are required for DNA replication, late gene expression and host cell shutoff (2). The largest protein encoded in the E4 region is E4orf6, which is required for productive adenovirus replication (1). E4orf6 forms a complex with another early gene product, E1B55k (3), and this complex associates with cellular proteins such as cullin 5, elongins B and C and Rbx, to form E3 ubiquitin ligase (4,5). This ligase targets p53 (4–7), the MRN complex (8), DNA ligase IV (9) and integrin α3 (10) for proteasomal degradation. This ubiquitin ligase activity is known to be required for the nuclear export of viral late mRNAs (11,12).
Several viral proteins encoded in the E4 region, such as E4orf1, E4orf3 and E4or6 are known to have oncogenic activities (13–17). E4orf1 of adenovirus type 9, which belongs to adenovirus subgroup D, has been shown to be involved in mammary tumorigenesis (14,18). E4orf3 and E4orf6 proteins of subgroup C adenovirus type 5 have the potential to transform cells in cooperation with E1A and E1B proteins and enhance the growth of tumors transplanted in nude mice (15–17). In a previous report, we demonstrated that in cells transformed with adenovirus type 5 E4orf6, cellular pp32 protein associates with E4orf6 and AU-rich element (ARE)-containing mRNAs are exported to the cytoplasm in a chromosome region maintenance 1 (CRM1)-independent manner (19). Furthermore, the exported ARE-mRNAs are stabilized and those mRNAs acquire the potential to transform cells (20).
AREs usually exist in the 3′-untranslated region (UTR) of certain mRNAs encoding early response genes or growth-related genes, such as proto-oncogenes and growth factors (21,22). AREs are targets for rapid degradation of mRNA (21,23) and the fate of ARE-mRNA is controlled by several RNA-binding proteins, such as AUF1, tristetraprolin (TTP) and HuR (24). HuR, which is a member of the embryonic lethal abnormal vision (ELAV) family of RNA-binding proteins, binds to AREs in order to protect ARE-mRNA from rapid degradation (23,24). HuR is able to shuttle between the nucleus and cytoplasm, whereas it is mainly located in the nucleus. HuR-mediated stabilization of ARE-mRNA depends on HuR localization in the cytoplasm (24,25). In normal cells, HuR transiently relocalizes to the cytoplasm under conditions of stress. On the other hands, HuR constitutively accumulates in the cytoplasm of cancer cells and the cytoplasmic expression of HuR is thought to be involved in malignant transformation of cancer cells (25,26).
These facts suggest that the E4orf6-deleted mutant adenovirus is able to proliferate selectively in cancer cells in which ARE-mRNA is stabilized. In the present study, we examined the oncolytic activity of the adenovirus E4orf6-deleted mutant, dl355. The ability of this virus to replicate was markedly increased in cancer cells compared with that in normal cells. dl355 showed cytolytic activity for cancer cells in vitro and in vivo. The propagation and cytolytic activity of this virus were higher than those of an E1B55k-deleted adenovirus. These findings indicate that dl355 is a potential oncolytic virus.
Materials and methods
Cell lines, viruses and antibodies
The human lung cancer cell lines, A549 and H1299; cervical carcinoma cell lines HeLa, HeLa S3 and C33A; African green monkey kidney (Vero) cells carrying an integrated copy of the Ad5 E4 region, W162; human embryonal kidney cell line 293 (transformed by the adenovirus E1 gene); human foreskin fibroblast cell line, BJ; and normal human lung primary cell line, WI38 were used in the present study. All cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Ardrich; Merck KGaA, Darmstadt, Germany) containing 10% fetal bovine serum (FBS; Biowest, Nuaille, France) with antibiotics at 37°C in a 5% CO2 atmosphere under humidified conditions.
Wild-type adenovirus type 5 (WT300), E4orf6-deleted mutant adenovirus (dl355) (generous gift from Dr T. Shenk; Princeton University) and E1B55k-deleted mutant adenovirus (dl1520) (generous gift from Dr A.J. Berk; University of California) were used in the present study.
A western blot analysis was performed as previously described (27) using antibodies specific to E1A (M73; generous gift from Dr T. Shenk, Princeton University), E4orf6 (RSA#3; generous gift from Dr T. Shenk; Princeton University), actin (dilution 1:1,000; cat. no. sc-1616; Santa Cruz Biotechnology, Dallas, TX, USA) and HuR (dilution 1:2,500; cat. no. sc-5261; Santa Cruz Biotechnology) and β-tubulin (dilution 1:1,000; cat. no. 05-661; EMD Millipore Corp., Darmstadt, Germany) as primary antibodies. The secondary antibody was horseradish peroxidase-conjugated anti-goat IgG (dilution 1:5,000; cat. no. 805-035-180; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and horseradish peroxidase-conjugated anti-mouse IgG (dilution 1:5,000; cat. no. 115-035-062; Jackson ImmunoResearch Laboratories). Antibody binding was visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Preparation of dl355 and dl1520 lysates
To prepare virus lysates, dl355-infected W162 cells and dl1520 or WT300-infected 293 cells were subjected to three cycles of freezing and thawing. Virus concentrations [virus particles (vp)/ml] were then determined by a QuickTiter Adenovirus Quantitation kit (Cell Biolabs, San Diego, CA, USA). Viral titers [infectious units (ifu)/ml] were determined using the Adeno-X™ Rapid Titer kit (Clontech Laboratories, Inc., Mountain View, CA, USA) according to the manufacturer's instructions. To use a virus in in vivo experiments, its extract was purified using a Fast-Trap Adenovirus Purification and Concentration kit (Millipore, Billerica, MA, USA) according to the manufacturer's protocols.
Cytopathic effect assay and cell viability assay
Human cancer and normal cells were plated on 24-well plates (5×104 cells/well). Twenty-four hours later, the cells were infected with dl355 at a multiplicity of infection (MOI) of 0.1, 0.5, 1, 10 or 100 vp/cell and maintained for an additional 7 days. Cells were then fixed and stained with Coomassie brilliant blue.
A 2-3-bis [2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt assay was used to examine cell metabolic activity. Cancer and normal cells were seeded on 96-well plates at a density of 3.0×103 cells/well. Twenty-four hours later, the cells were infected with dl355, dl1520 or WT300 at an MOI of 100 vp/cell. Cell metabolic activity was determined using an XTT assay on days 1, 3, 5 and 7 with the Cell Proliferation kit II (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's protocol.
In vitro virus proliferation assay
Cancer and normal cells were seeded at 5.0×104 cells/well 24 h before the infection. Cells were infected with dl355, dl1520, or WT300 at an MOI of 1 vp/cell. These cells were incubated at 37°C for 48 h, after which cells were collected and a virus lysate was prepared as described above. Viral titers (ifu/ml) were determined using the Adeno-X Rapid Titer kit (Clontech Laboratories).
HuR depletion
For RNA interference analysis, Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific) was used to transfect HeLa cells with 20 nM of each small interfering RNA (siRNA) targeting HuR (5′-TTCGTAAGTTATTTCCTTTAATT-3′) or with a negative control siRNA (5′-TCTTAATCGCGTATAAGGCTT-3′; Qiagen, Hilden, Germany). After 48 h of transfection, HeLa cells were infected with dl355. After 24 h of infection, all cells were collected and the virus lysate was prepared using three freeze-thaw cycles. Viral titers were determined using the Adeno-X Rapid Titer kit (Clontech Laboratories) and 293 cells.
For heat shock treatment, HeLa cells were incubated at 43°C for 2 h immediately after dl355 infection, and the infected cells were heat shocked (2 h) again at 24 h after infection. Cells were harvested at 28 h after infection and the viral titers were determined as described above.
In vivo human tumor model
Female BALB/c nu/nu mice (purchased from Hokudo, Sapporo, Japan) were housed under specific pathogen-free conditions. The temperature was 26–28°C, and the light/dark cycle was 10-h/14-h cycle, food and water were taken ad libitum. HeLa S3 cells (1.0×106 cells/mouse) were injected subcutaneously into the flanks of mice (5 week old and 20–24 g) and permitted to grow to ~5–6 mm in diameter. The mice were randomly divided into two groups (5 per group) and 109 vp (100 µl) of dl355 or the same volume of PBS was injected twice (days 0 and 3) directly into the tumors. The perpendicular diameters of the tumors were measured every 3 or 4 days and tumor volumes were calculated using the following equation: Volume (mm3) = A × B2 × 0.5 (A is the longest diameter, B is the shortest diameter). The mice were sacrificed by cervical dislocation after 30 days of injection of virus. All procedures performed in this study involving animals were in accordance with the ethical standards of the Animal Care and Use Committee of the Hokkaido University (Sapporo, Japan).
Results
Selective dl355 replication in cancer cells
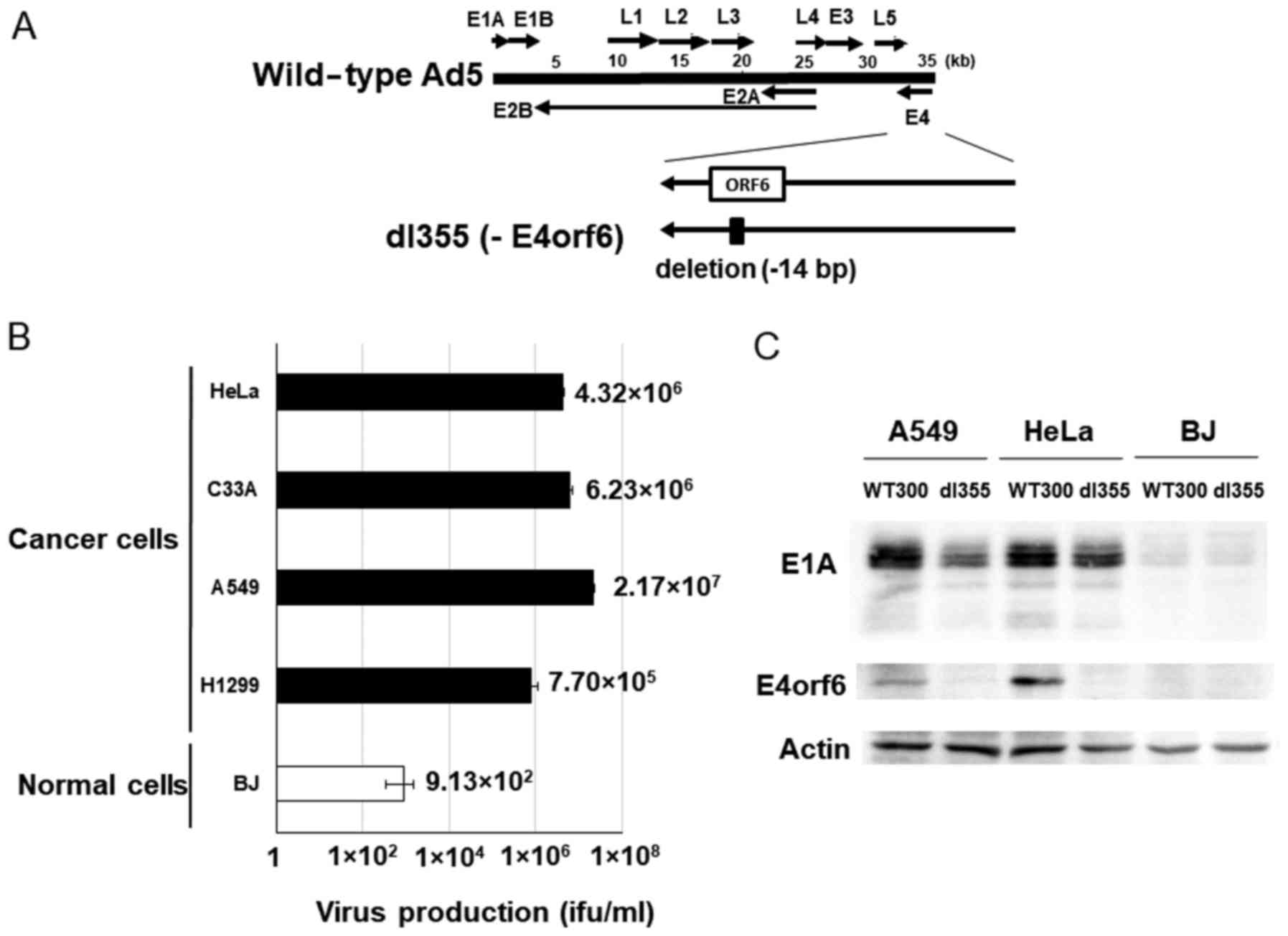
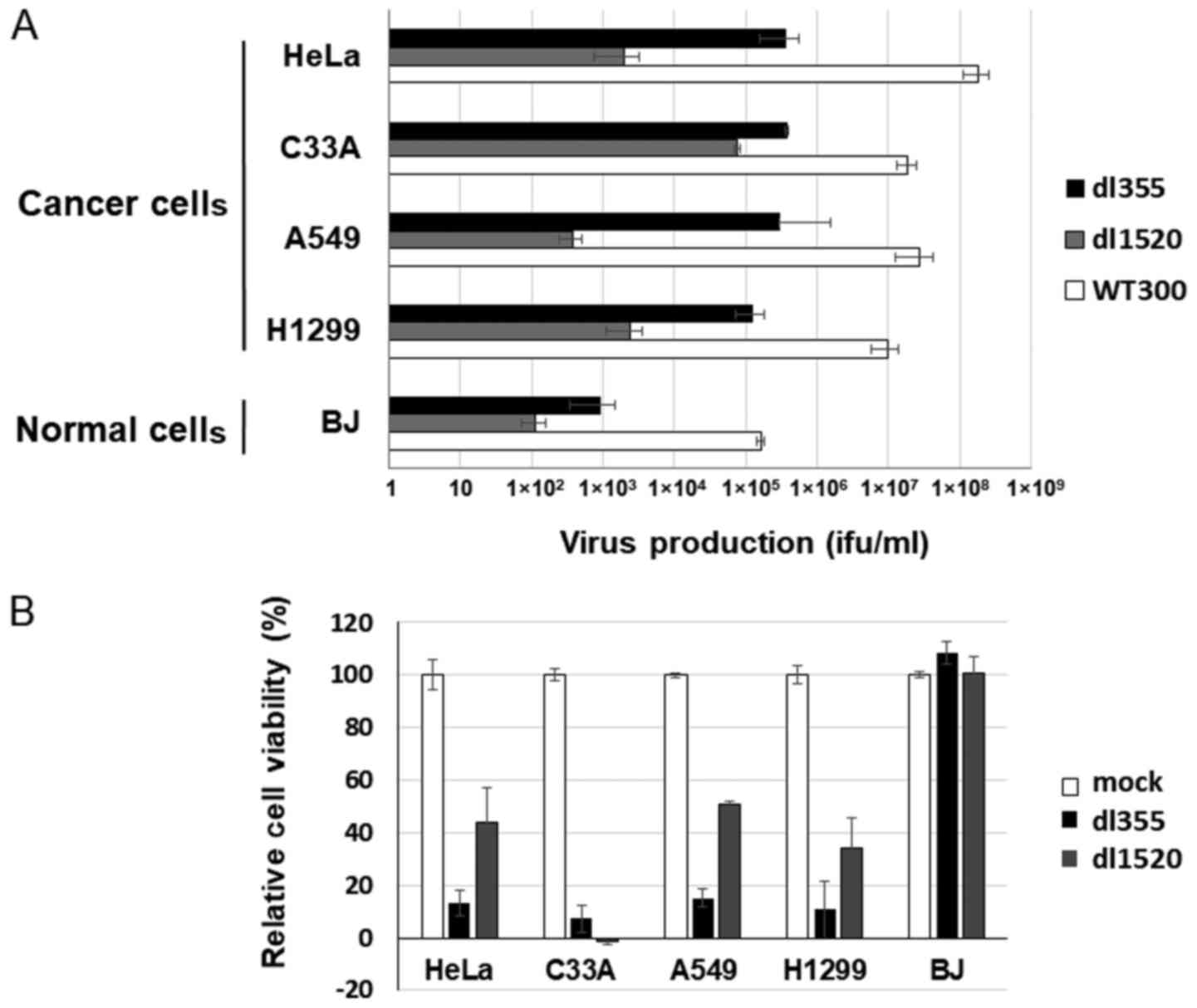
dl355 (Fig. 1A) has a 14-bp deletion in the E4orf6 gene and was constructed in 1985 to investigate the functions of genes located in the E4 region in adenovirus-infected host cells. This virus showed deficient virus DNA replication, accumulation of late viral mRNAs, and shutoff of host cell mRNAs compared to wild-type adenovirus type 5 (Ad5) (2). To examine the productive efficiency of dl355, cancer cells (HeLa, C33A, A549 and H1299) and normal cells (BJ) were infected with dl355 at an MOI of 1 virus particle (vp)/cell and virus titers generated after 48 h were detected by staining the hexon protein of virus particles in 293 cells. In these cancer cells, the propagation of dl355 was very high, with titers from 7.70×105 to 2.17×107 ifu/ml. On the other hand, the titer of dl355 in normal cells (BJ; foreskin fibroblasts) was 3 to 5 logs lower (9.13×102 ifu/ml) than in cancer cells (Fig. 1B). We examined the expression of E1A protein, which is expressed first after infection. The amount of E1A protein was at a high level in dl355-infected A549 and HeLa cells, although the level was low in normal BJ cells (Fig. 1C). These results suggest that dl355 is selectively produced in cancer cells.
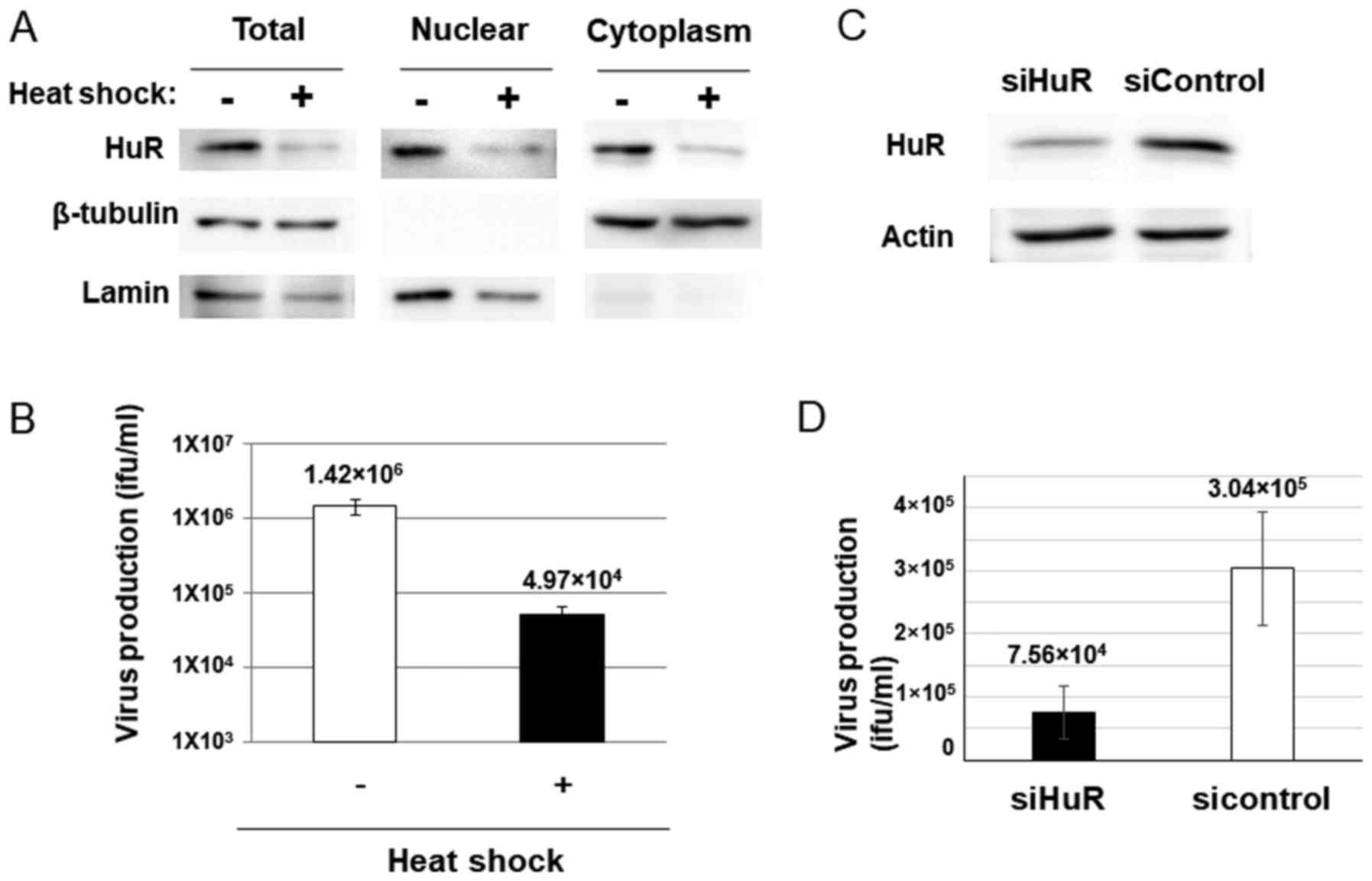
Since dl355 was thought to propagate in cancer cells in which ARE-mRNA is stabilized, we examined whether the ARE-mRNA stabilization system was required for dl355 replication. To evaluate this, we confirmed adenovirus production in HuR-depleted cells, because decreased HuR inhibits ARE-mRNA stabilization (28). Heat shock (HS) treatment is known to downregulate HuR by ubiquitin-mediated proteolysis and HuR-targeted mRNA is also decreased in HS-treated cells (29). If dl355 replicates using the ARE-mRNA stabilization system, the virus titer is expected to reduce with HS treatment. HeLa cells were subjected to HS, as described in Materials and methods, and were then examined for HuR protein and virus production. As expected, a 2-h HS treatment of HeLa cells resulted in reduced expression of HuR protein in the cytoplasm of HeLa cells (Fig. 2A). Furthermore, HS-treated cells showed a significant reduction in virus production compared to the non-treated cells (~1/28.6; Fig. 2B).
Since HS treatment affects have many influences other than HuR proteolysis in cells, we confirmed the HuR-depletion effect by HuR knockdown (KD). The virus production in siRNA for HuR introduced cells (Fig. 2C) was much less than that of the cells with control siRNA transfected cells (~1/4; Fig. 2D). These results suggest that HuR plays an important role in dl355 replication.
In vitro and in vivo cytolytic potential of dl355
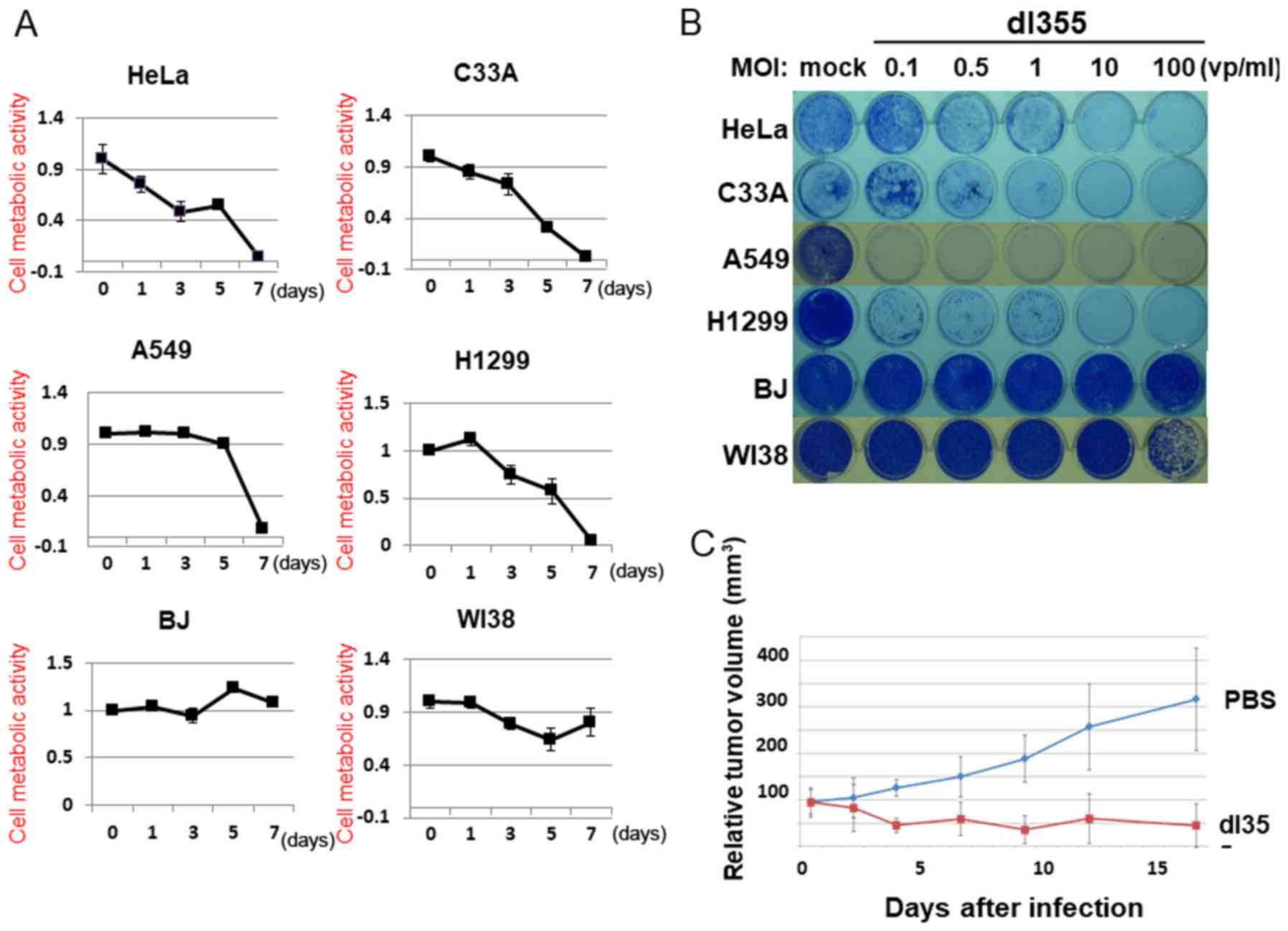
To estimate the cell lysis activity of dl355, we examined cell viability using the XTT assay. HeLa, A549, C33A, H1299, BJ and WI38 cells were infected with dl355 at an MOI of 100 (vp/cells) and the XTT assay was performed 1, 3, 5 and 7 days after infection (Fig. 3A). In the case of HeLa cells, dl355 infection resulted in reduced cell viability 3 days after infection and most cells died after 7 days. HeLa cells showed the earliest effects, but in other cancer cells, nearly all cells died on day 7. In contrast, most normal cells infected with dl355 did not die even after 7 days.
To assess the cell lysis activity of dl355 further, we examined the infected cells using a cytopathic effects (CPE) assay. Four different cancer cell types (HeLa, C33A, A549 and H1299) and two normal cell types (BJ and WI38) were infected with dl355 at MOIs of 0.1, 0.5, 1, 10 or 100. Cytotoxicity was estimated by staining the remaining cells with Coomassie brilliant blue 7 days after infection (Fig. 3B). Although several cell lines survived with low MOIs, all cancer cells were killed by dl355 in a dose-dependent manner. On the other hand, most normal cells survived in all infections. Taken together, these results demonstrate the in vitro-selective cytolytic activity of dl355.
We estimated the therapeutic effect of dl355 for human cancers using a tumor xenograft model. In order to examine this, HeLa S3 cells were implanted into the hind flanks of 5-week-old female BALB/c nu/nu mice. When the tumors grew to ~5–6 mm in diameter, 109 vp dl355 (100 µl) or the same volume of phosphate-buffered saline (PBS) (as a control) were injected twice (days 0 and 3) directly into the tumor. Tumor growth was significantly suppressed by injection with dl355 whereas the PBS-injected tumors grew almost three times as large in 18 days (Fig. 3C). Thus, dl355 exerted significant effects on human tumor xenografts in nude mice.
Comparison of the oncolytic effects of dl355 and dl1520
Since an E1B-55k gene deleted-adenovirus has been developed as an oncolytic adenovirus (30) and is already applied clinically, we next compared the oncolytic activity of dl355 with that of the E1B-55k-deleted adenovirus, dl1520. Using cancer cells, HeLa, C33A, A549 and H1299, and normal BJ cells, dl355, dl1520 and WT300 as a control of infectivity were infected at an MOI of 1 and virus titers were estimated 48 h after infection. Since different cell lines (293 and W162 cells) were used to determine the virus production rate (ifu/ml) of each virus, we compared the productive efficiencies of both viruses with viral particles (vp/ml). As shown in Fig. 4A, dl355 virus production was significantly higher (1–3 logs higher) than dl1520 virus production in all cancer cells.
To compare cell lysis activity, both viruses (MOI 100) were infected into the same cancer and normal cells and the XTT assay was performed 7 days after infection. Except for C33A case, dl355 showed stronger cell death activity than dl1520 in cancer cells (Fig. 4B). However, in normal cells, both viruses showed almost no cell death effect. Taken together, these data indicate that dl355 has a stronger oncolytic effect than dl1520 in at least several cancer cell lines.
Discussion
In the present study, we described the oncolytic potential of the E4orf6-deleted adenovirus, dl355. The productive efficiencies of this virus with cancer cells were approximately 102 to 103 times higher from that of normal cells (Fig. 1B). It showed a high level of cytolytic activity for cervical and lung cancer cells in vitro and the same effect was evident in a tumor xenograft model. Furthermore, dl355 has oncolytic activity that may be superior to dl1520, which is currently used clinically. These findings indicate that dl355 is a potential oncolytic virus.
Many types of conditionally replicative adenoviruses (CRAds) targeted to cancer cells are being developed and several viruses are currently in clinical trials (31). Oncolytic adenoviruses can be divided into at least two types (32), one of which has mutations in genes required for viral replication. For example, the E1A or E1B-55k gene deleted-virus has been developed as a CRAd and it has oncolytic effects for pRB- or p53-deficient tumor cells (30,33). The other group consists of viruses that possess a cancer-specific transcription system in the virus genes required for replication such as E1A. For example, promoters of the telomerase gene (34) or prostate-specific antigen gene (35) are inserted into the 5′-untranslated region (UTR) of the E1A gene to produce CRAds, which are specifically activated in cancer cells. There are few reports describing oncolytic viruses with tumor selectivity based on the level of mRNA stability. Thus, dl355 is a rare type of oncolytic virus that its replication is controlled by RNA modulation.
As we have shown in a previous study (28), HuR KD attenuates the export and stabilization of ARE-mRNA. This is due to the fact that HuR is the only protein that binds directly to ARE, so the ARE-mRNA failed to be exported to the cytoplasm if HuR disappears. In this study, virus proliferation was downregulated under HuR HS or HuR KD conditions. These results suggest that the export and stabilization of ARE-mRNA is essential for the growth of dl355. In a previous study (36), it was clarified that various stimuli promote the relocalization of HuR to the cytoplasm and the stabilization of ARE-mRNA. Therefore, if stimulation is added to enhance export and stabilization of ARE-mRNA, the production efficiency of dl355 increases, indicating the possibility of obtaining a stronger tumor lysis activity.
We used various types of cancer cells in this study and it is expected that the infection efficiency of the virus was different in each cell line. We examined the expression of E1A protein, which is expressed first after infection, to evaluate the infectivity of dl355. As the amount of E1A protein was almost the same between dl355-infected A549 and HeLa cells (Fig. 1C), the infectivity can be considered to be equivalent. Furthermore, as shown in Fig. 4A, the virus production of the WT300 infected in different types of cancer cells was not significantly different, thus the infection efficiency was not so different. These data showed the validity of comparing the production efficiency of dl355 and dl1520.
In the vast majority of cancer cells, ARE-mRNAs relocate to the cytoplasm with HuR and are constitutively stabilized (26). We showed that dl355 replication depends on the ARE-mRNA stabilization system, since the replication of dl355 is downregulated in HuR-depleted cancer cells (Fig. 2). These facts suggest that dl355 has the potential to be effective for many types of cancer cells.
Acknowledgements
The authors thank Dr T. Shenk (Princeton University) for providing dl355 and wild-type (WT300) adenoviruses and antibodies and Dr A.J. Berk (University of California) for providing the mutant adenovirus, dl1520, and the members of our laboratories for their helpful discussions and support.
Funding
The present study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan (nos. 26293423 and 23659928).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
FH, AYM and YM conceived and designed the research and contributed to the writing of the paper. AYM and YM mainly conducted the research. UH, TK and MTA conducted the in vivo analysis. MY and UH contributed to the production of the virus. FH, MY, YK, KM and MS assisted with statistical analysis and also analyzed the data. TK, YK and MS proofread the paper. KM and MTA revised the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the study are appropriately investigated and resolved.
Ethics approval and consent to participate
All the animal experiments performed in this study were in accordance with the ethical standards of the Animal Care and Use Committee of Hokkaido University (Sapporo, Hokkaido, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Shenk T: Adenoviridae: The viruses and their replication. Fundamental Virology. Knipe DM and Howley PM: 4th. Lippincott Williams & Wilkins Ltd.; Philadelphia: pp. 1053–1088. 2001 | |
|
Halbert DN, Cutt JR and Shenk T: Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 56:250–257. 1985.PubMed/NCBI | |
|
Sarnow P, Hearing P, Anderson CW, Halbert DN, Shenk T and Levine AJ: Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 49:692–700. 1984.PubMed/NCBI | |
|
Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW and Branton PE: Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Harada JN, Shevchenko A, Shevchenko A, Pallas DC and Berk AJ: Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 76:9194–9206. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Luo K, Ehrlich E, Xiao Z, Zhang W, Ketner G and Yu XF: Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 21:1742–1750. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng CY, Blanchette P and Branton PE: The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology. 364:36–44. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Stracker TH, Carson CT and Weitzman MD: Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 418:348–352. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Baker A, Rohleder KJ, Hanakahi LA and Ketner G: Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 81:7034–7040. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Dallaire F, Blanchette P, Groitl P, Dobner T and Branton PE: Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J Virol. 83:5329–5338. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Woo JL and Berk AJ: Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J Virol. 81:575–587. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Blanchette P, Kindsmüller K, Groitl P, Dallaire F, Speiseder T, Branton PE and Dobner T: Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J Virol. 82:2642–2651. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Täuber B and Dobner T: Adenovirus early E4 genes in viral oncogenesis. Oncogene. 20:7847–7854. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Javier RT: Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J Virol. 68:3917–3924. 1994.PubMed/NCBI | |
|
Nevels M, Täuber B, Kremmer E, Spruss T, Wolf H and Dobner T: Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol. 73:1591–1600. 1999.PubMed/NCBI | |
|
Moore M, Horikoshi N and Shenk T: Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 93:11295–11301. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Nevels M, Rubenwolf S, Spruss T, Wolf H and Dobner T: The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 94:1206–1211. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Javier R, Raska K Jr and Shenk T: Requirement for the adenovirus type 9 E4 region in production of mammary tumors. Science. 257:1267–1271. 1992. View Article : Google Scholar : PubMed/NCBI | |
|
Higashino F, Aoyagi M, Takahashi A, Ishino M, Taoka M, Isobe T, Kobayashi M, Totsuka Y, Kohgo T and Shindoh M: Adenovirus E4orf6 targets pp32/LANP to control the fate of ARE-containing mRNAs by perturbing the CRM1-dependent mechanism. J Cell Biol. 170:15–20. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Kuroshima T, Aoyagi M, Yasuda M, Kitamura T, Jehung JP, Ishikawa M, Kitagawa Y, Totsuka Y, Shindoh M and Higashino F: Viral-mediated stabilization of AU-rich element containing mRNA contributes to cell transformation. Oncogene. 30:2912–2920. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Chen CY and Shyu AB: AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 20:465–470. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Jacobson A and Peltz SW: Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 65:693–739. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Brennan CM and Steitz JA: HuR and mRNA stability. Cell Mol Life Sci. 58:266–277. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Hinman MN and Lou H: Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 65:3168–3181. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
López de Silanes I, Lal A and Gorospe M: HuR: Post-transcriptional paths to malignancy. RNA Biol. 2:11–13. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES and Gorospe M: Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 22:7146–7154. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Aoyagi M, Higashino F, Yasuda M, Takahashi A, Sawada Y, Totsuka Y, Kohgo T, Sano H, Kobayashi M and Shindoh M: Nuclear export of adenovirus E4orf6 protein is necessary for its ability to antagonize apoptotic activity of BH3-only proteins. Oncogene. 22:6919–6927. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Kakuguchi W, Kitamura T, Kuroshima T, Ishikawa M, Kitagawa Y, Totsuka Y, Shindoh M and Higashino F: HuR knockdown changes the oncogenic potential of oral cancer cells. Mol Cancer Res. 8:520–528. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, et al: Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28:1271–1282. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, et al: An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 274:373–376. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
Larson C, Oronsky B, Scicinski J, Fanger GR, Stirn M, Oronsky A and Reid TR: Going viral: A review of replication-selective oncolytic adenoviruses. Oncotarget. 6:19976–19989. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Bressy C and Benihoud K: Association of oncolytic adenoviruses with chemotherapies: An overview and future directions. Biochem Pharmacol. 90:97–106. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A, Hawkins L and Kirn D: An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 6:1134–1139. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Kawashima T, Kagawa S, Kobayashi N, Shirakiya Y, Umeoka T, Teraishi F, Taki M, Kyo S, Tanaka N and Fujiwara T: Telomerase-specific replication-selective virotherapy for human cancer. Clin Cancer Res. 10:285–292. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Rodriguez R, Schuur ER, Lim HY, Henderson GA, Simons JW and Henderson DR: Prostate attenuated replication competent adenovirus (ARCA) CN706: A selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 57:2559–2563. 1997.PubMed/NCBI | |
|
Wang J, Guo Y, Chu H, Guan Y, Bi J and Wang B: Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 14:10015–10041. 2013. View Article : Google Scholar : PubMed/NCBI |