MicroRNA‑936 inhibits the malignant phenotype of retinoblastoma by directly targeting HDAC9 and deactivating the PI3K/AKT pathway
Retraction in: /10.3892/or.2022.8389
- Authors:
- Published online on: January 9, 2020 https://doi.org/10.3892/or.2020.7456
- Pages: 635-645
-
Copyright: © Xu et al. This is an open access article distributed under the terms of Creative Commons Attribution License [CC BY_NC 4.0].
Abstract
Introduction
Retinoblastoma (RB) is a type of human malignant tumor originating from the primitive retinal layer (1). This disease has familial transmissibility and usually occurs in children under 3 years of age (2). It is estimated that RB-related deaths account for approximately 1% of all deaths among infants and young children (3). Currently, treatment strategies for patients with RB include chemotherapy, ophthalmectomy, laser therapy and cryotherapy (4). In recent years, tremendous advances in therapeutic techniques have greatly improved the clinical outcomes of patients with RB; unfortunately, the long-term survival rates are still extremely unsatisfactory due to invasion and metastasis (5,6). RB can directly infiltrate into the brain via the optic nerve or invade other tissues via the blood stream (7). The genesis and progression of RB are very complex processes, which involve gene mutation, activation of oncogenes, and inactivation of tumor suppressors; however, the detailed molecular events have not been fully documented to date. Hence, in-depth exploration of the mechanisms behind RB initiation and progression is urgently necessary for identifying novel therapeutic approaches and improving clinical outcomes.
MicroRNAs (miRNAs) are a group of endogenous non-coding short RNA molecules that have the capacity to regulate the expression of their target genes (8). They negatively modulate the expression of target genes through directly interacting with the 3′-untranslated regions of mRNA, thus causing either mRNA degradation or translation repression (9). In total, 4,469 genes of miRNAs, including 1,881 precursor and 2,588 mature miRNAs, have been identified in the human genome, according to miRBase. In the field of RB research, a variety of different miRNAs have been revealed to be aberrantly expressed, and their anomalous expression plays important roles in the regulation of multiple cancer-related processes, including cell survival, proliferation, apoptosis, metastasis, angiogenesis and epithelial-mesenchymal transition (10). For instance, miR-98 (11), miR-186 (12) and miR-665 (13) are weakly expressed in RB and restrain tumor progression; on the contrary, miR-93 (14), miR-106b (15) and miR-198 are overexpressed in RB and promote tumor aggressiveness.
miR-936 has been reported to exert important effects on the progression of non-small cell lung cancer (16) and glioma (17). However, the expression and functions of miR-936 in RB remain elusive and need to be further elucidated. In our study, we aimed to detect miR-936 expression in RB, to evaluate its clinical significance, and identify the functional importance in the oncogenicity of RB. Moreover, the mechanisms behind the tumor-suppressive activity of miR-936 in RB cells in vitro and in vivo were investigated in detail. The findings of our study will offer novel insights into the pathogenesis of RB and may facilitate the identification of new targets for anticancer therapies.
Materials and methods
Human tissue samples
This study was conducted with the approval of the Ethics Committee of West China Hospital (2016.0407) and was carried out following the guidelines of the Declaration of Helsinki. In addition, informed consent forms were signed by all the participants. A total of 33 RB tissue samples were obtained from patients with RB who had not been treated with preoperative radiotherapy, chemotherapy or other anticancer modalities. Normal retinal tissues were collected from the ruptured globes of 12 patients. All the patients (mean age, 21 years old; age range, 16–47 years) underwent ophthalmectomy at West China Hospital between May 2016 to February 2018. After surgical resection, all tissues were snap-frozen in liquid nitrogen and then transferred to a −80°C cryogenic refrigerator.
Cell culture
Three RB cell lines, Y79, Weri-RB1, and SO-RB50 as well as a human normal retinal pigmented epithelium cell line APRE-19 were purchased from the American Type Culture Collection (ATCC). Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% of fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% of a penicillin/streptomycin solution (Gibco; Thermo Fisher Scientific, Inc.) was used for cell culture. All cells were grown at 37°C in a humidified atmosphere supplied with 5% of CO2.
Transfection assay
The miR-936 agomir (agomir-936) and negative control (NC) agomir (agomir-NC) were generated by Shanghai GenePharma Co., Ltd.. The agomir-936 sequence was 5′-ACAGUAGAGGGAGGAAUCGCAG-3′ and the agomir-NC sequence was 5′-UUGUACUACACAAAAGUACUG-3′. Small interfering (si)RNA directed against the human HDAC9 mRNA (si-HDAC9) and the NC siRNA (si-NC) were chemically synthesized by Guangzhou RiboBio Co., Ltd. An HDAC9 overexpression plasmid lacking its 3′ untranslated region (3′-UTR), pcDNA3.1-HDAC9 (pc-HDAC9), and the empty pcDNA3.1 plasmid were purchased from GeneChem. Cells in the logarithmic growth phase were seeded in 6-well plates. After overnight incubation, the agomir (50 nM), siRNA (100 pmol) or plasmid (4 µg) were introduced into the cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The transfected cells were used in the subsequent experiments.
Reverse-transcription quantitative PCR (RT-qPCR)
Expression of miR-936 and HDAC9 mRNA was determined via RT-qPCR analysis. In particular, the isolation of total RNA from tissues or cells was conducted by means of TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was then subjected to reverse transcription for cDNA synthesis with the miScript Reverse Transcription Kit (Qiagen GmbH). After that, the miScript SYBR Green PCR Kit (Qiagen GmbH) was utilized to detect miR-936 expression. To quantify HDAC9 mRNA expression, cDNA was reverse-transcribed from total RNA using the PrimeScript™ RT Reagent Kit (Takara Biotechnology, Co., Ltd.). Next, cDNA was amplified using the SYBR-Green PCR Master Mix (Takara Biotechnology, Co., Ltd.). U6 small nuclear RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) respectively served as endogenous controls for miR-936 and HDAC9 mRNA expression. Relative gene expression was calculated by the 2−ΔΔCq method (18).
The primers were designed as follows: miR-936, 5′-CACGCAACAGTAGAGGGA-3′ (forward) and 5′-CCAGTGCAGGGTCCGAGGTA-3′ (reverse); U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); HDAC9, 5′-ATGGTTTCACAGCAACGCATT-3′ (forward) and 5′-ACCTTGCCTAAGCGTCTGC-3′ (reverse); and GAPDH, 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward) and 5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse).
Cell Counting Kit-8 (CCK8) and colony formation assays
Transfected cells were harvested and seeded in 96-well plates at a density of 2×103 cells per well. Cellular proliferation was analyzed by the addition of 10 µl of the CCK-8 solution (Beyotime Institute of Biotechnology) into each well. The absorbance was measured on a microplate reader (Molecular Devices). The CCK-8 assay was conducted at four time points: 0, 24, 48 and 72 h after seeding.
A colony formation assay was performed for evaluating the colony-forming ability of RB cells. A total of 500 transfected cells were seeded in 6-well plates and then incubated at 37°C for 2 weeks. At the end of this assay, colonies were fixed with 4% paraformaldehyde, stained with methyl violet, and washed thrice with phosphate-buffered saline (PBS; Gibco; Thermo Fisher Scientific, Inc.). The colonies were counted under an inverted light microscope (Olympus X71; Olympus Corp.).
Flow cytometric analysis of apoptosis
The proportion of apoptotic cells was examined with the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Biolegend). Transfected cells (~1.5×106) were collected, washed twice with ice-cold PBS, and resuspended in 100 µl of binding buffer. After the addition of Annexin V and propidium iodide (PI) solutions, the cell suspension was incubated at room temperature for 15 min in darkness and then transferred to a flow cytometer tube. Finally, the early (Annexin V-FITC+/PI−) + late (Annexin V-FITC+/PI+) apoptosis rate was measured on a flow cytometer (FACScan™; BD Biosciences).
Transwell migration and invasion assays
The 24-well Transwell chambers (8-µm pore size) coated with Matrigel (both from BD Biosciences) were used to assess cellular invasiveness. Transected cells were collected at 48 h post-transfection, centrifuged, and resuspended in FBS-free DMEM medium. A total of 200 µl of the cell suspension containing 5×104 cells was added into the top chambers. The lower chambers were covered with 500 µl of the culture medium containing 20% of FBS. After 24 h, the noninvasive cells that remained in the top chambers were gently wiped off. The invasive cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Following extensive washing, the invasive cells were imaged by means of an inverted light microscope (magnification, ×200). The invasive cells that migrated through the pores were counted for the determination of cell invasion. The Transwell migration assay was carried out similarly to the Transwell invasion assay, except that the 24-well Transwell chambers were not precoated with Matrigel.
Subcutaneous heterotopic xenograft assay
All the experimental procedures involving animals were approved by the Animal Care and Use Committee of West China Hospital (2016.0802) and were carried out in compliance with the Animal Protection Law of the People's Republic of China-2009 for experimental animals. Four- to six-week-old female BALB/c nude mice (20 g) were obtained from Shanghai Laboratory Animal Center (Shanghai, China). The animals were maintained under specific pathogen-free conditions (25°C, 50% humidity, 10-h light/14-h dark cycle) and ad libitum food/water access. Hetero-transplantation was conducted through subcutaneous inoculation of agomir-936- or agomir-NC-transfected Y79 cells (1×107) into the flanks of nude mice (n=4 for each group). The width and length of xenografts that formed were measured at 2-day intervals with a caliper. The allowable maximum tumor size is 2.0 cm. All the nude mice were euthanized by means of cervical dislocation 4 weeks after the implantation. The tumor xenografts were excised, weighed, and stored for further use. Tumor volumes were calculated via the following formula: Tumor volume = length × (width)2/2.
Bioinformatic analysis and the luciferase reporter gene assay
Two miRNA target prediction and functional study databases, TargetScan7.1 (http://www.targetscan.org/) and miRDB (http://mirdb.org/), were employed to search for the potential targets of miR-936.
The wild-type (WT) 3′-UTR of the HDAC9 mRNA containing the predicted miR-936-binding site and a mutant (MUT) HDAC9 3′-UTR were synthesized by Shanghai GenePharma Co., Ltd. The WT and MUT 3′-UTR fragments were inserted into the pMIR-REPORT luciferase reporter plasmid (Ambion; Thermo Fisher Scientific, Inc.), resulting in the HDAC9-WT and HDAC9-MUT reporter plasmids. Cells were seeded in 24-well plates and serum-starved for 6 h before the transfection. Cotransfection of either the HDAC9-WT or HDAC9-MUT reporter plasmid and either agomir-936 or agomir-NC was performed with Lipofectamine® 2000 reagent. After cultivation for 48 h, the transfected cells were harvested and subjected to the quantification of luciferase activity using a Dual-Luciferase Reporter Assay System (Promega, Corporation). Renilla luciferase activity served as an internal reference.
Western blot analysis
Cells or homogenized tissue samples were lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) containing a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Total protein was quantified by means of the BCA Kit (Beyotime Institute of Biotechnology). Equal amounts of total protein (20 µg) were loaded and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis on a 10% gel, followed by electroblotting onto polyvinylidene fluoride membranes and by blocking at room temperature with 5% non-fat milk for 2 h. Subsequent to overnight incubation at 4°C, the corresponding secondary antibodies (cat no. ab205719 and ab6721, dilution 1:5,000; Abcam) were applied to probe the membranes. After three washes with Tris-buffered saline containing 0.1% of Tween-20 (TBST), an Enhanced Chemiluminescence Detection System (Pierce; Thermo Fisher Scientific, Inc.) was added, and the protein signals were visualized after 1 min. Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.) was utilized for densitometry.
The following primary antibodies were employed: anti-HDAC9 (molecular weight: 111 kDa; cat. no. ab109446, dilution 1:1,000, rabbit monoclonal antibody; Abcam), anti-p-PI3K p85 alpha (phospho Y607; molecular weight: 84 kDa; cat no. ab182651, dilution 1:1,000, rabbit monoclonal antibody; Abcam), anti-PI3K (molecular weight: 85 kDa; cat. no. ab86714, dilution 1:1,000, mouse monoclonal antibody; Abcam), anti-p-AKT (Ser 473 phosphorylated Akt1, Ser 474 phosphorylated Akt2 and correspondingly Ser 472 phosporylated Akt3; molecular weight: 60 kDa; cat. no. sc-81433, dilution 1:1,000, mouse monoclonal antibody; Santa Cruz Biotechnology), anti-AKT (molecular weight: 62 kDa; cat. no. sc-56878, dilution 1:1,000, mouse monoclonal antibody; Santa Cruz Biotechnology) and anti-GAPDH (molecular weight: 37 kDa; cat. no. sc-51907, dilution 1:1,000, mouse monoclonal antibody; Santa Cruz Biotechnology).
Statistical analysis
All experiments were repeated at least three times, and statistical analyses were performed using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). The association between miR-936 expression and clinical parameters of RB patients was assessed by the χ2 test. Student's t-test was carried out for comparisons of two groups, whereas the differences among multiple groups were evaluated by one-way ANOVA followed by Tukey's post hoc test. The expression correlation between miR-936 and HDAC9 mRNA was investigated by Spearman's correlation analysis. All data are shown as mean ± standard deviation, and statistically significant differences were defined as those with P<0.05.
Results
miR-936 expression is low in RB tissue samples and cell lines
To clarify the miR-936 expression profile in RB, RT-qPCR analysis was carried out to determine miR-936 expression in 33 RB tissue samples and 12 normal retinal tissue samples. The expression of miR-936 was weak in the RB tissue samples compared with that noted in the normal retinal tissues (Fig. 1A, P<0.05). In addition, miR-936 expression was determined in three RB cell lines: Y79, Weri-RB1, and SO-RB50. A human normal retinal pigmented epithelium cell line, APRE-19, served as the control. The results of RT-qPCR analysis indicated that all three RB cell lines relatively underexpressed miR-936 compared relative to APRE-19 cells (Fig. 1B, P<0.05).
We next explored the correlation between miR-936 expression and the clinical parameters among the patients with RB. All subjects were subdivided into miR-936 low- or miR-936 high-expression groups according to the median value of miR-936 among the RB tissue samples. As indicated in Table I, low miR-936 expression notably correlated with patient tumor differentiation (P=0.037), lymph node metastasis (P=0.010) and TNM staging (P=0.005). These results implied that downregulated miR-936 in RB may correlate with cancer progression.
miR-936 exerts inhibitory action on the growth and metastasis of RB cells in vitro
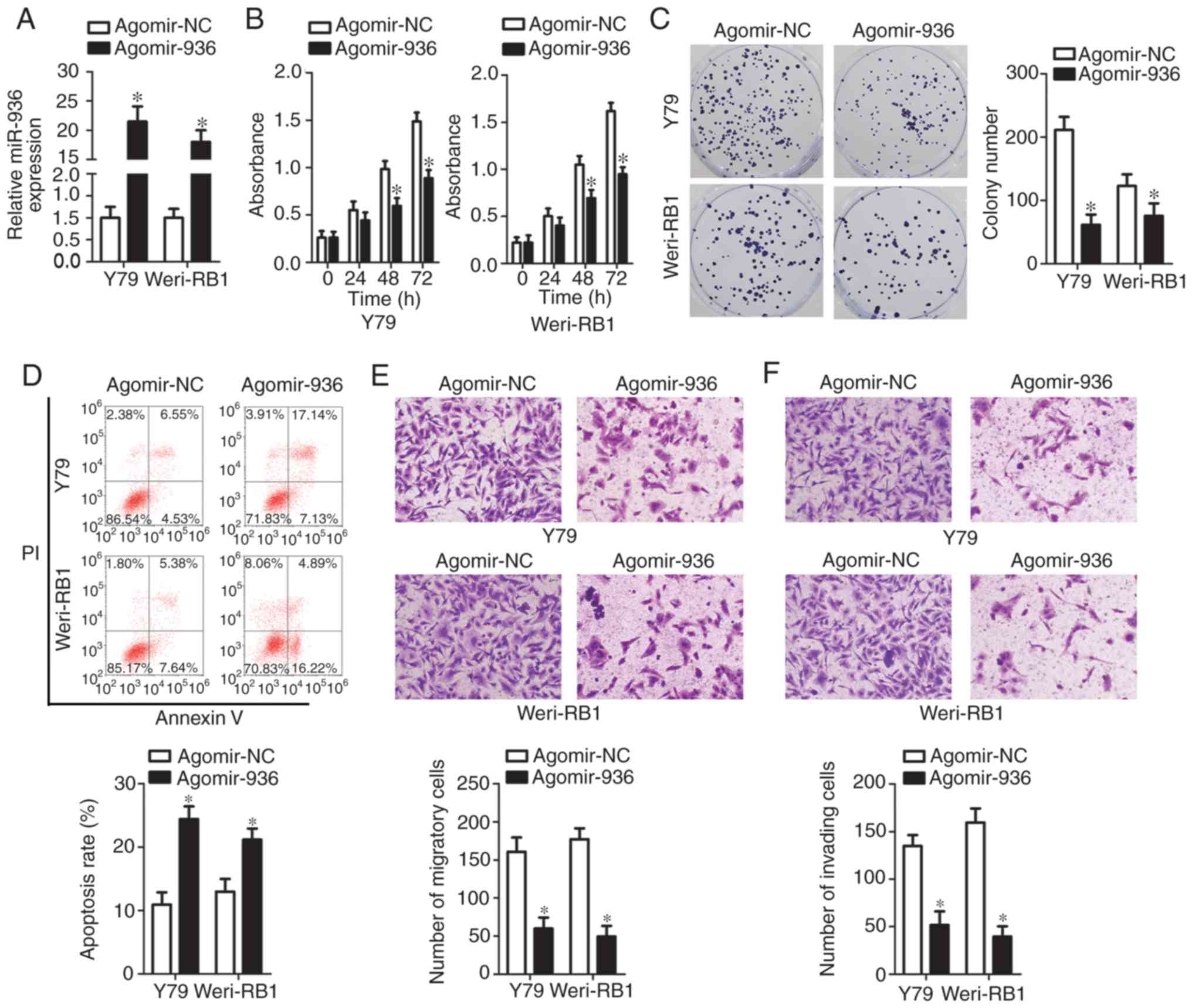
Cell lines Y79 and Weri-RB1 manifested relative lower miR-936 expression among the three tested RB cell lines; hence, the two cell lines were chosen for subsequent experiments. To investigate the detailed involvement of miR-936 in the malignancy of RB, Y79 and Weri-RB1 cells were treated with agomir-936 or agomir-NC. Transfection of agomir-936 notably increased endogenous miR-936 expression in the Y79 and Weri-RB1 cells relative to the cells transfected with agomir-NC (Fig. 2A, P<0.05). The CCK-8 assay revealed that the recovery of miR-936 expression significantly impeded the proliferation of Y79 and Weri-RB1 cells (Fig. 2B, P<0.05). The results of the colony formation assay were similar to the results of the CCK-8 assay; the colonies in the agomir-936 groups were fewer and smaller than those in the agomir-NC groups (Fig. 2C, P<0.05).
A significant inhibition of RB cell proliferation in response to miR-936 upregulation prompted us to test whether miR-936 can affect apoptosis. To this end, flow cytometric analysis was performed to measure the apoptosis rate of Y79 and Weri-RB1 cells after agomir-936 or agomir-NC transfection. Ectopic miR-936 expression significantly promoted the apoptosis of Y79 and Weri-RB1 cells (Fig. 2D, P<0.05). We next conducted Transwell migration and invasion assays to detect the influence of miR-936 overexpression on the migratory and invasive abilities of RB cells. Transfection of agomir-936 resulted in an obvious decrease in the numbers of migratory (Fig. 2E, P<0.05) and invading (Fig. 2F, P<0.05) Y79 and Weri-RB1 cells. Collectively, our data revealed that miR-936 overexpression inhibited proliferation and migration, while it promoted apoptosis, in RB cells in vitro.
Histone deacetylase 9 (HDAC9) is a direct target gene of miR-936 in RB cells
To elucidate the mechanism of action of miR-936 in the regulation of the aggressiveness of RB cells, a potential target of miR-936 was next predicted through bioinformatic analysis (Fig. 3A): it was determined to be HDAC9. The luciferase reporter assay was performed to test whether miR-936 could directly bind to the 3′-UTR of HDAC9 mRNA. The data indicated that compared with the agomir-NC group, exogenous miR-936 expression notably decreased the luciferase activity of HDAC9-WT (Fig. 3B, P<0.05); however, the luciferase activity of the HDAC9-MUT was unaffected in Y79 and Weri-RB1 cells after agomir-936 introduction. Next, we measured HDAC9 expression in miR-936-overexpressing Y79 and Weri-RB1 cells by RT-qPCR and western blotting. The HDAC9 mRNA expression was decreased by agomir-936 transfection in Y79 and Weri-RB1 cells (Fig. 3C, P<0.05). Consistently with this finding, the protein amount of HDAC9 manifested the same alteration in Y79 and Weri-RB1 cells (Fig. 3D, P<0.05). Furthermore, we determined the mRNA level of HDAC9 in 33 RB tissue samples and 12 normal retinal tissue samples by RT-qPCR analysis. It was observed that HDAC9 mRNA was upregulated in RB tissues compared with that in normal retinal tissues (Fig. 3E, P<0.05). Furthermore, a negative expression correlation between miR-936 and HDAC9 mRNA was identified among our 33 clinical RB tissue samples (Fig. 3F; R2=0.3336, P=0.0004). Taken together, these results indicated that HDAC9 is a direct target gene of miR-936 in RB cells.
Downregulation of HDAC9 imitates the effects of miR-936 overexpression in RB cells
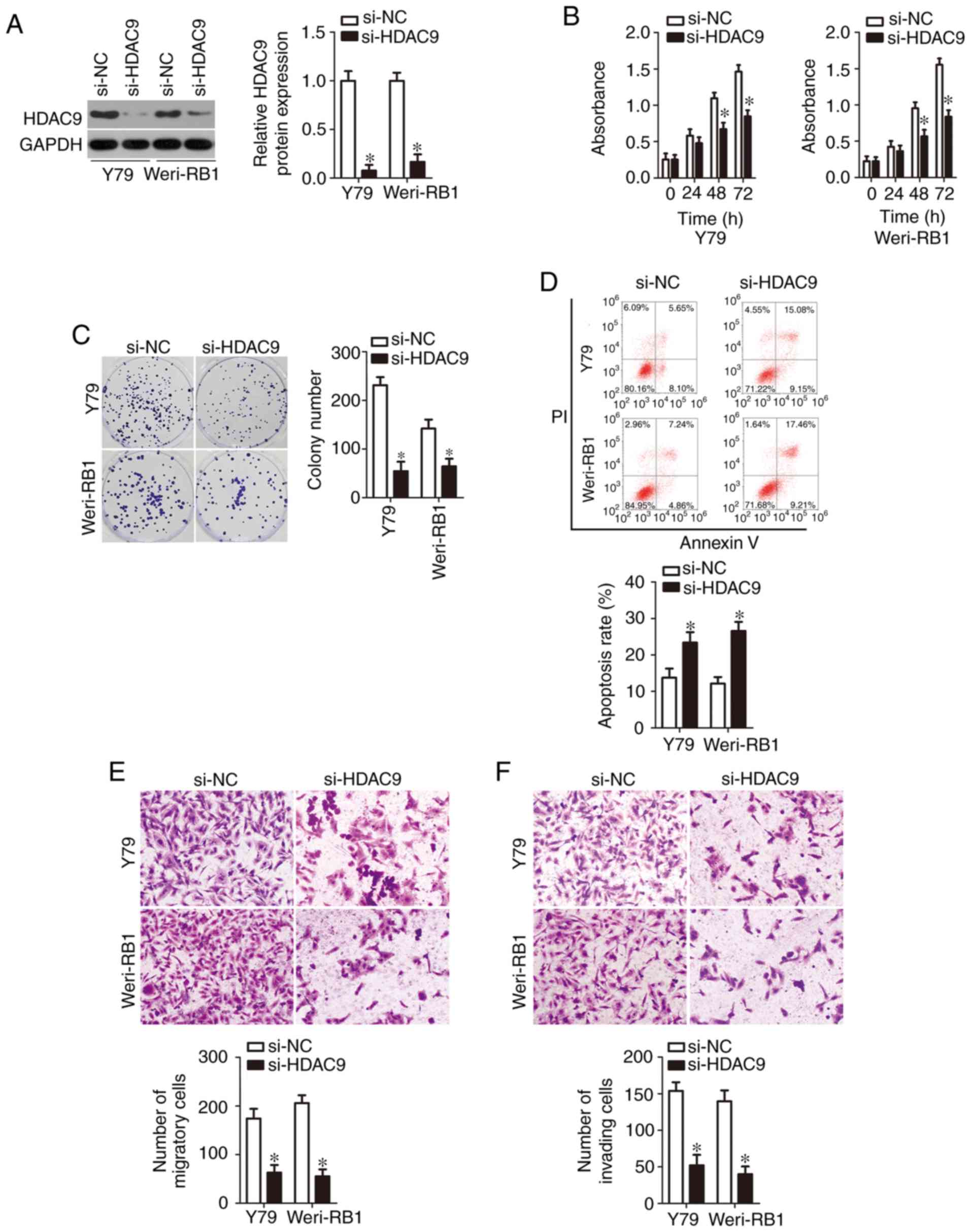
Having identified HDAC9 mRNA as a direct target of miR-936, we next explored the biological roles of HDAC9 in RB cells. A loss-of-function experiment was conducted on Y79 and Weri-RB1 cells after si-HDAC9 or si-NC transfection. Transfection efficiency was confirmed by western blotting and is depicted in Fig. 4A (P<0.05). Next, a series of functional experiments was performed, and the results suggested that silencing of HDAC9 expression significantly inhibited Y79 and Weri-RB1 cell proliferation (Fig. 4B, P<0.05) and colony formation (Fig. 4C, P<0.05); promoted apoptosis (Fig. 4D, P<0.05); and impaired cell migration (Fig. 4E, P<0.05) and invasion (Fig. 4F, P<0.05) in vitro. These observations revealed that the functional effects of HDAC9 knockdown were similar to those caused by restoration of miR-936 expression in RB cells, thereby confirming HDAC9 mRNA as a functional target of miR-936.
Recovery of HDAC9 expression neutralizes the action of miR-936 overexpression in RB cells
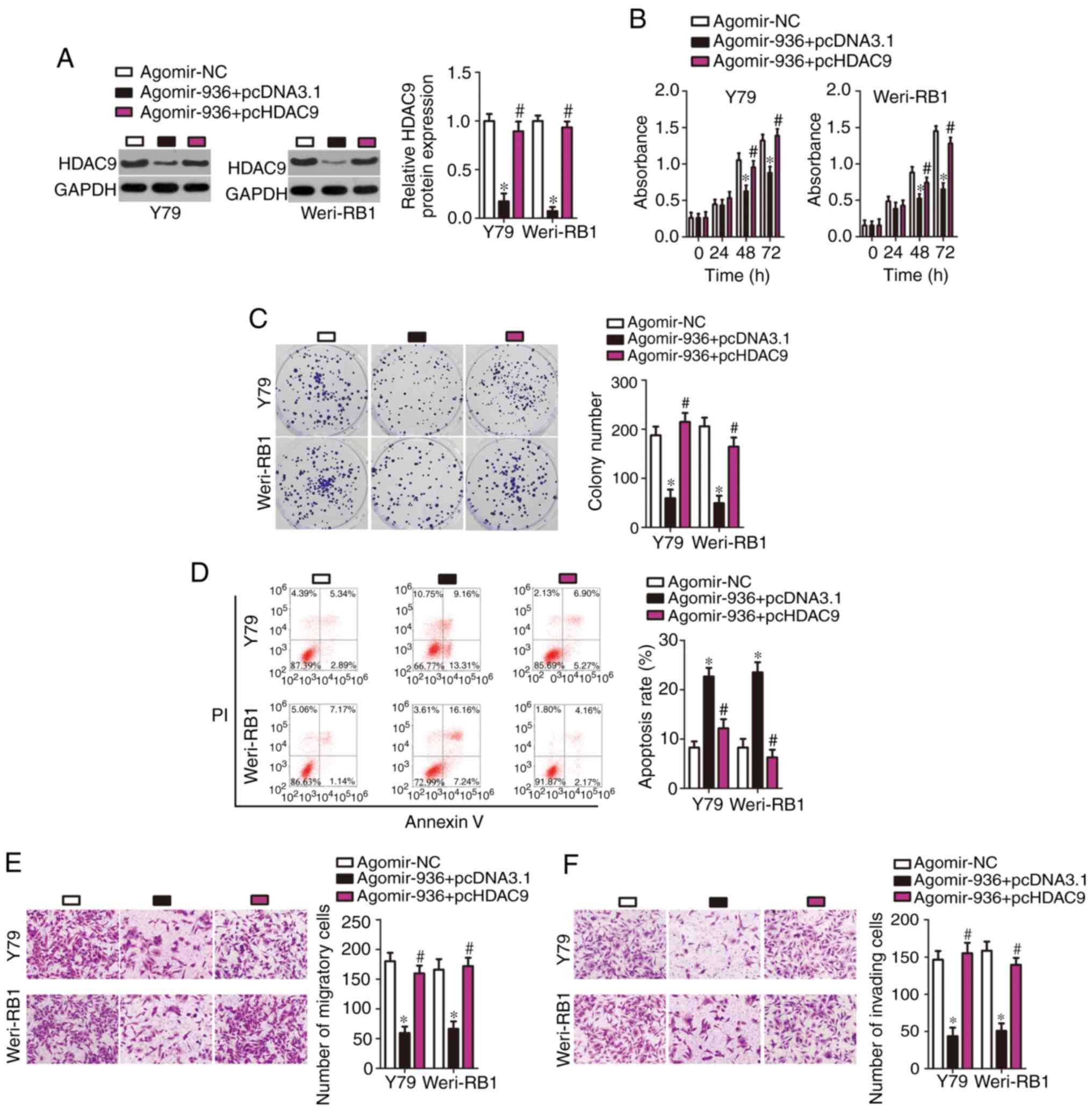
To clarify whether miR-936-dependent inhibition of the malignant characteristics of RB is mediated directly by HDAC9, rescue experiments were performed by introducing an HDAC9 overexpression plasmid lacking its 3′-UTR pc-HDAC9 or empty pcDNA3.1 plasmid into miR-936-overexpressing Y79 and Weri-RB1 cells. The results of western blotting indicated that the transfection of agomir-936 resulted in an obvious decrease in HDAC9 protein expression in Y79 and Weri-RB1 cells, whereas cotransfection of pc-HDAC9 abrogated the suppressive effects of miR-936 on HDAC9 protein expression (Fig. 5A, P<0.05). Then, additional experiments on cell proliferation, colony formation, apoptosis, migration and invasion were carried out, and the results revealed that the miR-936-mediated effects on Y79 and Weri-RB1 cell proliferation (Fig. 5B, P<0.05), colony formation (Fig. 5C, P<0.05), apoptosis (Fig. 5D, P<0.05), migration (Fig. 5E, P<0.05), and invasion (Fig. 5F, P<0.05) were substantially reversed after reintroduction of HDAC9. Again, these results revealed that HDAC9 downregulation mediated the modulatory influence of miR-936 on the malignant phenotype of RB cells.
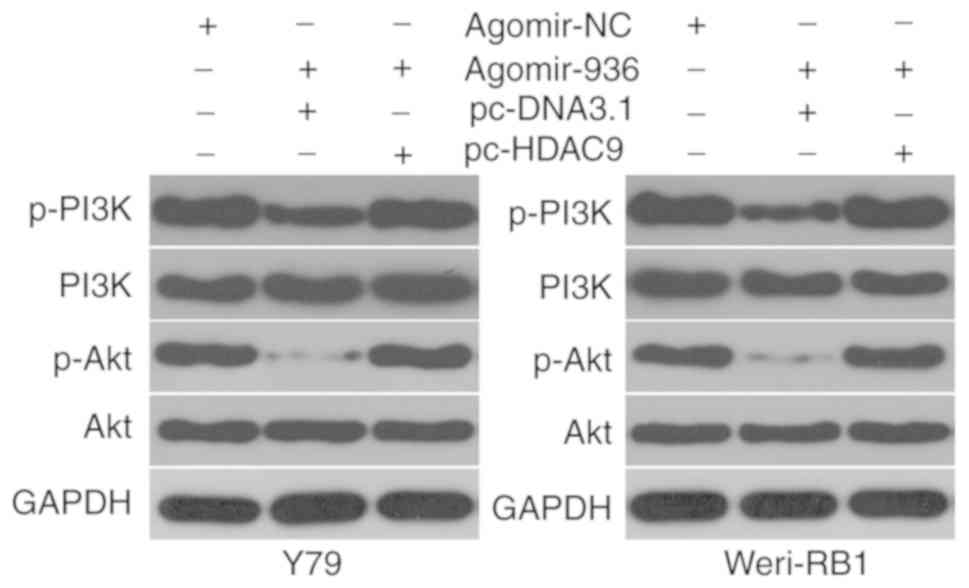
miR-936 deactivates the PI3K/AKT pathway in RB cells by targeting HDAC9
Previous research suggested that HDAC9 is involved in activation of the PI3K/AKT signaling pathway (19). Whether or not miR-936 inhibits HDAC9-mediated activation of PI3K/AKT signaling in RB cells was examined by cotransfection of Y79 and Weri-RB1 cells with agomir-936 and either pcDNA3.1 or pc-HDAC9, and then the molecules associated with the PI3K/Akt pathway were determined by western blotting. miR-936 overexpression significantly reduced the protein amounts of p-PI3K and p-AKT in Y79 and Weri-RB1 cells; however, the levels of total PI3K and AKT stayed unaltered. Notably, the restored HDAC9 expression counteracted the inhibitory effects of miR-936 upregulation on the cellular levels of p-PI3K and p-AKT (Fig. 6). These results suggest that miR-936 inhibits activation of the PI3K/AKT pathway in RB cells by directly targeting HDAC9 mRNA.
miR-936 impairs RB xenograft growth in vivo
Although we validated the direct inhibitory effect of miR-936 upregulation on RB cell proliferation in vitro, it was necessary to test the influence of miR-936 on tumor growth in vivo. Hence, the subcutaneous heterotopic xenograft experiment was conducted by injection of agomir-936- or agomir-NC-transfected Y79 cells into the flanks of nude mice. The nude mice inoculated with agomir-936-transfected Y79 cells featured slower tumor growth (Fig. 7A and B, P<0.05) and smaller tumor weight (Fig. 7C, P<0.05). Finally, tumor xenografts were obtained for RT-qPCR and western blotting. Compared with the agomir-NC group, the expression of miR-936 was obviously higher (Fig. 7D, P<0.05), whereas HDAC9, p-PI3K, and p-Akt protein amounts were lower (Fig. 7E), in the tumor xenografts derived from the agomir-936 group. These results revealed that miR-936 overexpression decreased tumor growth in vivo by inhibiting the HDAC9/PI3K/AKT pathway.
Discussion
Several lines of evidence have revealed that various aberrantly expressed miRNAs exist in RB, and multiple genes directly targeted by these miRNA are implicated in the regulation of RB pathogenesis (20–22). Almost all aggressive phenotypes of RB have been continuously reported to be regulated by miRNAs. Moreover, miRNAs have the potential to be validated as possible diagnostic and prognostic biomarkers, as well as promising targets for anticancer therapies (23). Accordingly, a comprehensive study of the cancer-related miRNAs in RB may help to identify more effective therapeutic interventions, thereby improving the prognosis for patients with this disease. Whether miR-936 may be useful in RB gene therapy is yet to be determined. In this study, we measured miR-936 expression in RB tissue samples and cell lines, assessed its clinical value among patients with RB, investigated the detailed effects of miR-936 on RB progression and elucidated its mechanism of action. Our results provide a novel insight into the network of miR-936/HDAC9/PI3K/AKT signaling in RB.
miR-936 is weakly expressed in non-small cell lung cancer (16) and glioma (17). Patients with glioma harboring lower miR-936 expression have shorter overall survival than patients with higher miR-936 expression (17). Functionally, miR-936 has been identified as a tumor-suppressive miRNA in the above-mentioned two human cancer types. Ectopic miR-936 expression was found to decrease non-small cell lung cancer cell proliferation and invasion but to promote apoptosis in vitro. Recovery of miR-936 expression was found to hinder tumor growth of glioma in vitro and in vivo (17). Nevertheless, few studies have illustrated the expression status and detailed roles of miR-936 in RB. Herein, our results revealed that miR-936 expression is low in both RB tissue samples and cell lines. The decreased miR-936 expression was significantly associated with differentiation, lymph node metastasis and TNM staging among patients with RB. Further experiments indicated that upregulation of miR-936 restricted RB cell proliferation and the colony formation ability of these cells, promoted their apoptosis, and attenuated cell migration and invasion in vitro. Further investigation revealed that miR-936 overexpression delayed RB tumor growth in vivo, indicating that miR-936 inhibited the tumorigenesis of RB in vivo. These observations suggest that miR-936 may be an effective target for the anticancer therapy of RB.
Validation of the molecular mechanisms underlying the tumor-suppressive action of miR-936 on the malignancy of RB may be helpful for exploring effective therapeutic strategies. Previously, E2F2 (16) and CKS1 (17) have been demonstrated to be the direct target genes of miR-936. In the present study, the mechanisms related to miR-936-mediated restriction of the aggressive phenotype of RB cells in vitro and in vivo were explored at the molecular level. First, bioinformatic analysis predicted that the 3′-UTR of HDAC9 matches the seed sequence of miR-936. Second, luciferase reporter assays confirmed that miR-936 can directly bind to the 3′-UTR of HDAC9 mRNA. Third, miR-936 overexpression decreased HDAC9 expression in RB cells at the mRNA and protein levels. Fourth, HDAC9 was found to be overexpressed in RB tissues, manifesting an inverse expression correlation with miR-936. Fifth, a reduction in HDAC9 expression imitated the effects of miR-936 upregulation in RB cells. Finally, restoration of HDAC9 expression counteracted the tumor-suppressive influence of miR-936 on the oncogenicity of RB cells. These observations collectively identified HDAC9 as a direct target of miR-936 in RB cells.
HDAC9 is a member of the histone deacetylase (HDAC) family, which is implicated in multiple biological phenomena, including transcriptional regulation and cell death, particularly in carcinogenesis and cancer progression (24–26). In RB alone, HDAC9 is upregulated, and its upregulation obviously correlates with regional lymph node classification, largest tumor base, and tumor differentiation (27). Patients with RB harboring high HDAC9 expression show shorter overall survival and progression-free survival than do the patients with low HDAC9 expression (27). HDAC9 has been proposed to work as an oncogene in RB initiation and progression by affecting a variety of pathophysiological processes (27,28). HDAC9 is able to decrease EGFR expression and thereby inhibits the downstream PI3K/AKT signaling pathway activation, resulting in facilitating cancer progression (19); accordingly, we also tested whether miR-936 was implication in the control of the PI3K/AKT pathway in RB cells by regulating HDAC9. The results of our present study indicate that miR-936 directly targets HDAC9 to inhibit the activation of the PI3K/AKT pathway, resulting in the inhibition of RB tumor growth. Therefore, HDAC9 silencing via miR-936 restoration may be an effective therapeutic modality for patients with RB.
Four limitations are included in the present study. Firstly, endogenous miR-936 was not silenced and subsequently the impacts of miR-936 knockdown in RB cells were not investigated. Secondly, the sample size was small. Third, a previous study reported a novel method to overexpress miRNAs in target cells both in vitro and in vivo through adeno-associated virus (29); according, further studies using this novel technique to increase miR-936 expression should be conducted, and this would be able to further demonstrate the tumor-suppressive activities of miR-936 in the malignancy of RB. Lastly, determination of the effect when miR-936 is antagonized in normal retinal pigmented epithelium cell line APRE-19 was not examined. Further investigations may resolve these issues.
To summarize, we demonstrated the tumor-suppressive properties of miR-936 in RB and reported for the first time that it may function by directly targeting HDAC9 mRNA and deactivating the PI3K/AKT pathway. The present study offers an innovative perspective on therapeutic approaches to RB.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Foundation of Nanchong City and University Cooperation Project (no. 18SXHZ0515) and the Foundation of Sichuan Provincial Education Department (no. 17ZA01711).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JZ and LX conceived and designed the study. LX, WL, QS, MW, HL, XY and JZ performed the experiments. JZ, LX and WL wrote the paper. LX, WL, QS, MW, HL, XY and JZ reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and informed consent
This study was conducted with the approval of the Ethics Committee of West China Hospital and was carried out following the guidelines of the Declaration of Helsinki. In addition, informed consent forms were signed by all the participants. All the experimental procedures involving animals were approved by the Animal Care and Use Committee of West China Hospital and were carried out in compliance with the Animal Protection Law of the People's Republic of China-2009 for experimental animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS and Gallie BL: Retinoblastoma. Lancet. 379:1436–1446. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Kivelä T: The epidemiological challenge of the most frequent eye cancer: Retinoblastoma, an issue of birth and death. Br J Ophthalmol. 93:1129–1131. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
He MY, An Y, Gao YJ, Qian XW, Li G and Qian J: Screening of RB1 gene mutations in Chinese patients with retinoblastoma and preliminary exploration of genotype-phenotype correlations. Mol Vis. 20:545–552. 2014.PubMed/NCBI | |
|
Balmer A, Zografos L and Munier F: Diagnosis and current management of retinoblastoma. Oncogene. 25:5341–5349. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Tian T, Ji XD, Zhang Q, Peng J and Zhao PQ: A delayed diagnosis of unsuspected retinoblastoma in an in vitro fertilisation infant with retinopathy of prematurity. Int J Ophthalmol. 9:1361–1363. 2016.PubMed/NCBI | |
|
Abramson DH, Shields CL, Munier FL and Chantada GL: Treatment of retinoblastoma in 2015: Agreement and disagreement. JAMA Ophthalmol. 133:1341–1347. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Correa-Acosta A, González-Alviar ME and Gaviria-Bravo ML: Retinoblastoma and optic nerve enhancement in a brain magnetic resonance scan: Is it always a metastasis? Arch Soc Esp Oftalmol. 93:251–254. 2018.(In English, Spanish). View Article : Google Scholar : PubMed/NCBI | |
|
Lytle JR, Yario TA and Steitz JA: Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 104:9667–9672. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Guarnieri DJ and DiLeone RJ: MicroRNAs: A new class of gene regulators. Ann Med. 40:197–208. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Delsin LEA, Salomao KB, Pezuk JA and Brassesco MS: Expression profiles and prognostic value of miRNAs in retinoblastoma. J Cancer Res Clin Oncol. 145:1–10. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Li W, Wang J, Zhang D, Zhang X, Xu J and Zhao L: MicroRNA-98 targets HMGA2 to inhibit the development of retinoblastoma through mediating Wnt/β-catenin pathway. Cancer Biomark. 25:79–88. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wu S, Han M and Zhang C: Overexpression of microRNA-186 inhibits angiogenesis in retinoblastoma via the Hedgehog signaling pathway by targeting ATAD2. J Cell Physiol. 234:19059–19072. 2019.PubMed/NCBI | |
|
Wang S, Du S, Lv Y, Zhang F and Wang W: MicroRNA-665 inhibits the oncogenicity of retinoblastoma by directly targeting high-mobility group box 1 and inactivating the Wnt/β-catenin pathway. Cancer Manag Res. 11:3111–3123. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Cao Y, Xia F, Wang P and Gao M: MicroRNA-93-5p promotes the progression of human retinoblastoma by regulating the PTEN/PI3K/AKT signaling pathway. Mol Med Rep. 18:5807–5814. 2018.PubMed/NCBI | |
|
Bu W, Wang Y and Min X: MicroRNA-106b promotes the proliferation, migration and invasion of retinoblastoma cells by inhibiting the expression of ZBTB4 protein. Exp Ther Med. 16:4537–4545. 2018.PubMed/NCBI | |
|
Zhou X and Tao H: Overexpression of microRNA-936 suppresses non-small cell lung cancer cell proliferation and invasion via targeting E2F2. Exp Ther Med. 16:2696–2702. 2018.PubMed/NCBI | |
|
Wang D, Zhi T, Xu X, Bao Z, Fan L, Li Z, Ji J and Liu N: MicroRNA-936 induces cell cycle arrest and inhibits glioma cell proliferation by targeting CKS1. Am J Cancer Res. 7:2131–2143. 2017.PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Yang R, Wu Y, Wang M, Sun Z, Zou J, Zhang Y and Cui H: HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget. 6:7644–7656. 2015.PubMed/NCBI | |
|
Li J, Zhang Y, Wang X and Zhao R: microRNA-497 overexpression decreases proliferation, migration and invasion of human retinoblastoma cells via targeting vascular endothelial growth factor A. Oncol Lett. 13:5021–5027. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Liang Y, Chen X and Liang Z: MicroRNA-320 regulates autophagy in retinoblastoma by targeting hypoxia inducible factor-1α. Exp Ther Med. 14:2367–2372. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Li J and You X: MicroRNA-758 inhibits malignant progression of retinoblastoma by directly targeting PAX6. Oncol Rep. 40:1777–1786. 2018.PubMed/NCBI | |
|
Golabchi K, Soleimani-Jelodar R, Aghadoost N, Momeni F, Moridikia A, Nahand JS, Masoudifar A, Razmjoo H and Mirzaei H: MicroRNAs in retinoblastoma: Potential diagnostic and therapeutic biomarkers. J Cell Physiol. 233:3016–3023. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Singh A, Patel P, Patel VK, Jain DK, Veerasamy R, Sharma PC and Rajak H: Histone deacetylase inhibitors for the treatment of colorectal cancer: Recent progress and future prospects. Curr Cancer Drug Targets. 17:456–466. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Dokmanovic M and Marks PA: Prospects: Histone deacetylase inhibitors. J Cell Biochem. 96:293–304. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Marks PA, Richon VM, Kelly WK, Chiao JH and Miller T: Histone deacetylase inhibitors: Development as cancer therapy. Novartis Found Symp. 259:269–281; discussion 281–288. 2004.PubMed/NCBI | |
|
Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H and Huang Z: Downregulation of HDAC9 inhibits cell proliferation and tumor formation by inducing cell cycle arrest in retinoblastoma. Biochem Biophys Res Commun. 473:600–606. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Jin Q, He W, Chen L, Yang Y, Shi K and You Z: MicroRNA-101-3p inhibits proliferation in retinoblastoma cells by targeting EZH2 and HDAC9. Exp Ther Med. 16:1663–1670. 2018.PubMed/NCBI | |
|
Li M, Tang Y, Wu L, Mo F, Wang X, Li H, Qi R, Zhang H, Srivastava A and Ling C: The hepatocyte-specific HNF4α/miR-122 pathway contributes to iron overload-mediated hepatic inflammation. Blood. 130:1041–1051. 2017. View Article : Google Scholar : PubMed/NCBI |