Rab25 suppresses colon cancer cell invasion through upregulating claudin‑7 expression
- Authors:
- Published online on: December 18, 2023 https://doi.org/10.3892/or.2023.8685
- Article Number: 26
-
Copyright: © Cho et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Rab25 is a member of the RAS-oncogene superfamily of small GTPase and implicated in cancer cell invasion and metastasis of various types of cancer (1–4). This small GTPase is exclusively expressed in epithelial cells and arbitrates recycling of proteins from the late endosome to the plasma membrane to maintain cellular polarity and cell signaling (5). Emerging evidence indicates the context-dependent characteristics of Rab25 in cancer cell progression. While Rab25 promotes cancer cell invasion in ER-positive breast, ovarian and gastric cancers (1,6,7), Rab25 is under expressed in colon and triple negative breast cancer and suppresses invasion and metastasis of these types of cancer (8,9). In addition, the level of Rab25 expression is inversely associated with colorectal patient survival, reinforcing the tumor suppressive role of Rab25 in colon cancer (8).
Claudins are major components of tight junctions and maintain cellular polarity. Disruption of claudins is associated with tumorigenesis. Among 27 known members of the claudin family, claudin-7 is distributed in both apical and basolateral membranes of epithelial cells and tissues (10). In addition, this tight junction protein is critical for maintaining epithelial cell-matrix communications and intestinal equilibrium (10). A plethora of studies suggest the pivotal role of claudin-7 in suppressing colon cancer progression. Claudin-7 expression is downregulated in various types of cancer including colorectal cancer (11–16). In addition, low expression of claduin-7 leads to poor outcome of colon cancer patients (12–14). Mechanistically, claudin-7 is known to co-localize with β1 integrin and loss of claudin-7 downregulates β1 integrin expression, which leads to lung cancer cell invasion (15). Furthermore, claudin-7 suppresses colon cancer cell tumorigenesis in a Rab25-dependent manner (16).
Previously, we showed that Rab25 induces cancer cell endothelial-mesenchymal transition (EMT) and invasiveness through the β1 integrin/EGFR/vascular endothelial growth factor (VEGF)1/Snail signaling cascades (17). In addition, Snail mediates Rab25-induced aggressiveness in various types of cancer cell (17,18). However, the molecular mechanism by which Rab25 suppresses colon cancer EMT and invasion has not been identified. The present study demonstrated, for the first time to the best of the authors' knowledge, that Rab25 suppressed colon cancer cell invasion by upregulating claudin-7 expression. Rab25 inhibited colon cancer cell EMT and invasion. In addition, Rab25 inactivated the EGFR/Ras/Snail signaling axis. Furthermore, Rab25 induced claudin-7 protein expression to suppress colon cancer cell invasion, providing novel biomarkers for colon cancer.
Materials and methods
Reagents
(3,4,5-dimethylathiazol-2yl)-5-diphenyl-tetrazolium bromide (MTT), chloroquine (CQ), cycloheximide (CHX) and MG132 were purchased from Millipore Sigma. All other reagents were of the purest grade available.
Cell culture
HCT-116 (CCL-247) cells were purchased from American Type Culture Collection. Caco-2 cells were obtained from Korean Cell Line Bank. HCT-116 and Caco-2 cells were cultured in RPMI-1640 and Minimal Essential Medium (MEM) (Cytiva), respectively, supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Cytiva).
Plasmid transfection
Transfection was performed as previously described (16). The cells were cultured to ~80% confluency in a 35 mm dish and were transiently transfected using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Briefly, 500 µl of Opti-MEM I (Gibco; Thermo Fisher Scientific, Inc.), Lipofectamine (5 µl), DNA (2 µg), scrambled short interfering (si)RNA (1 µl) and siRNA (10 µl) were used for transfection. The mixtures were sequentially incubated at room temperature (RT) for 15 min, added to the cultured cells and then incubated at 37°C for 24 h. The Rab25 cDNA was subcloned into a pcDNA3 vector and an empty pcDNA3 vector was used as a negative control. RAS-V12 constructs were kindly provided by Dr A.R Moon (Duksung University, Seoul). Claudin-7 constructs were purchased from OriGene Technologies, Inc. siRNAs against Rab25 (SASI_Hs01_00216284), claudin-7 #1 (SASI_Hs01_00214821), claudin-7 #2 (SASI_Hs01_00214822) were purchased from Millipore Sigma. The sequence of siRNAs was as follows: Rab25, 5′-GAGCCAUCACCUCGGCGUA-3′ (sense) and 5′-UACGCCGAGGUGAUGGCUC-3′ (antisense); claudin-7 #1, 5′-CUAUGCGGGUGACAACAUC-3′ (sense) and 5′-GAUGUUGUCACCCGCAUAG-3′ (antisense); claudin-7 #2, 5′-CUGGUAUGGCCAUCAGAUU-3′ (sense) and 5′-AAUCUGAUGGCCAUACCAG-3′ (antisense). Scrambled siRNA as a negative control was obtained from Invitrogen (cat. no. 12935112; Invitrogen; Thermo Fisher Scientific, Inc.).
Cell viability
Cell viability was performed using the MTT assay as previously described (19,20). The cells (3×104 cells/well) were cultured to ~80% confluency in a 24-well plate and were transfected for 24 h. Following two PBS washes, 0.5 mg/ml of MTT solution was added to the wells and incubated at 37°C for 2 h. Then, 1 ml of dimethyl sulfoxide (DMSO) was added to each well. Immediately after the purple formazan crystals dissolved, the solution was collected and pipetted into a 96-well plate. Optical density was measured using an ELISA plate reader (BioTek Instruments, Inc.) at 540 nm.
Reverse transcription-quantitative (RT-q) PCR
RT-qPCR was performed as previously described (21). The cells (1×105 cells/well) were cultured to ~80% confluency in a 35 mm dish and were transfected for 24 h. Total RNA was extracted from the cells using TRIzol® (Thermo Fisher Scientific, Inc.) and isolated by adding chloroform according to the manufacturer's protocols. Then, RNA was precipitated by mixing with isopropanol. The RNA pellet was solubilized in RNase-free water. The total RNA was reverse transcribed using dNTPs (Thermo Fisher Scientific, Inc.), oligo (dT) and M-MLV reverse transcriptase (Promega Corporation). The cDNA complex was synthesized using a TaKaRa PCR Thermal Cycler Dice® (Takara Bio, Inc.) according to the manufacturer's protocols. The cDNA complex was amplified using an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) according to the iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.) protocols with the following primer sets: KRAS, 5′-TGTTCACAAAGGTTTTGTCTCC-3′ (forward) and 5′-CCTTATAATAGTTTCCATTGCCTTG-3′ (reverse); CLDN1, 5′-TTTACTCCTATGCCGGCGAC-3′ (forward) and 5′-GAGGATGCCAACCACCATCA-3′ (reverse); CLDN7, 5′-AGTTAGGAGCCTTGATGCCG-3′ (forward) and 5′-GCACAGGGAGTAGGATACGC-3′ (reverse); RAB25, 5′-CCATCACCTCGGCGTACTAT-3′ (forward) and 5′-TTTGTTACCCACGAGCATGA-3′ (reverse); and β-actin, 5′-AGAGCTACGAGCTGCCTGAC-3′ (forward) and 5′-AGCATCGTGTTGGCGTACAG-3′ (reverse). The mixture was initially denatured at 95°C for 3 min and then performed 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 10 sec, and extension at 72°C for 30 sec. The β-actin gene was used as a control for calculating the ΔCq value. The RT-qPCR data were analyzed using the 2−ΔΔCq method (22). The results are from the experiment in triplicate of three independent experiments.
Immunoblotting
Immunoblotting was performed as previously described (23). The cells were lysed using RIPA buffer (Millipore Sigma) containing a protease inhibitor cocktail (Roche Applied Science). Protein concentrations were measured using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Samples (30 µg) were separated by 8% [E-cadherin, phosphorylated (p-)EGFR, EGFR] or 12% (Rab25, Snail, Slug, Twist, Ras, β-actin, claudin-1, claudin-7, K-RAS) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes. After blocking with EzBlockChemi (ATTO Corporation) at RT for 30 min, the membrane was treated with primary antibody at 4°C overnight. The E-cadherin antibody (cat. no. 610182) was purchased from BD Biosciences. Antibodies for Rab25 (cat. no. 4314), Snail (cat. no. 3879), Slug (cat. no. 9585), Twist (cat. no. 46702), Ras (cat. no. 3965), phosphorylated (p-)EGFR (cat. no. v3777) and EGFR (cat. no. 2085) were obtained from Cell Signaling Technology, Inc. Antibodies for β-actin (cat. no. 47778), claudin-1 (cat. no. sc-166338) and claudin-7 (cat. no. sc-17670) were purchased from Santa Cruz Biotechnology, Inc. The K-Ras antibody (cat. no. ab180772) was purchased form Abcam. All primary antibodies were used at 1:1,000 dilution. Then, the membrane was incubated with the secondary antibody at RT for 2 h. Anti-rabbit (cat. no. 31463) and anti-mouse (cat. no. 31437) secondary antibodies were purchased from Thermo Fisher Scientific, Inc. (1:3,000), while an anti-goat (cat. no. sc-2354) secondary antibody was purchased from Santa Cruz Biotechnology, Inc. (1:3,000). The band was visualized using ECL reagents (cat. no. RPN2232; Amersham; Cytiva), and band densitometry was measured using ImageJ 1.53 version (National Institutes of Health).
Immunofluorescence
Immunofluorescence was performed as previously described (24). The cells were fixed with cold methanol for 10 min and blocked with 1% bovine serum albumin (BSA; Rocky Mountain Biologicals, Inc.) solution. Briefly, antibodies of E-cadherin (cat. no. 610182; 1:500; BD Biosciences), Snail (cat. no. sc-271977; 1:500; Santa Cruz Biotechnology, Inc.) and claudin-7 (cat. no. ab207300; 1:500; Abcam) were used. The cells were reacted with Cy2-conjugated goat anti-mouse IgG (cat. no. 111-223-003; 1:500; Jackson ImmunoResearch Laboratories, Inc.) and Cy3-conjugated goat anti-rabbit IgG (cat. no. 111-156-003; 1:500; Jackson ImmunoResearch Laboratories, Inc.). The cell nuclei were stained with 4′,6′-diamidino-2-phenylindole dihydrochloride (cat. no. v62249; 1:1,000; Thermo Fisher Scientific, Inc.) at RT for 5 min. Fluorescence images were captured using confocal microscopy (LSM710 Carl Zeiss AG).
Wound healing assay
Wound healing assay was performed as previously reported (25). The cells were cultured to ~80% confluency in a 35 mm dish and were transfected at 37°C for 24 h. The cells were scraped using a 200 µl micropipette tip and placed in new serum-free medium. Images were captured immediately after scraping and after 24 and 36 h in the same locations. The wound closure rate was calculated by measuring the area of the wound at each time using ImageJ 1.53 version (National Institutes of Health), and the mean value of triple replicated experiments was shown.
In vitro migration and invasion assay
The in vitro migration assay was performed in triplicates using a 48-well microchemotaxis chamber as previously described (21). Briefly, trypsinized cells were resuspended at a density of 2×106 cells/ml in RPMI-1640. FBS medium (1%) was added to each well of the lower chamber. Type 1 collagen (Cell matrix Type I-P; Nitta Gelatin Inc.)-coated 8 µm (for HCT-116) or 12 µm (for Caco-2) pore polyvinyl pyrrolidine-free polycarbonate filters (Neuro Probe Inc.) were added for in vitro migration analysis. After incubation for 6 h at 37°C, invaded cells were fixed and stained with Diff-Quik reagents (Dade Behring Inc.). The average numbers of three random fields under the light microscope (magnification, ×200) of invasion filters were counted in each experiment.
The in vitro invasion assay was performed in triplicates using a 48-well microchemotaxis chamber as previously described (26). First, 1% FBS medium was added to each well of the lower chamber. Matrigel (Corning, Inc.), which contains extracellular matrix components, was used to laminate 8 µm (for HCT-116) or 12 µm (for Caco-2) pore polyvinyl pyrrolidine-free polycarbonate filters for in vitro invasion assay. Filters were laminated at RT for 1 h with Matrigel. The transfected cell suspension, 2×106 cells/ml, was added to each well of the upper chamber. Following incubation for 16–18 h at 37°C, invaded cells were fixed and stained with Diff-Quik reagents (Dade Behring Inc.). The average numbers of three random fields under the light microscope (magnification, ×200) of invasion filters were counted in each experiment.
Three-dimensional (3D) Matrigel invasion assay
The 3D Matrigel invasion assay was performed as previously described (27–30). Cancer cells were labeled with DiI (Thermo Fisher Scientific, Inc.). The mixture was prepared by mixing of 20% type I collagen and Matrigel and solidify in the 3 µm pore size Transwell inserts (Corning, Inc.). A total of 5×104 cells were mixed in 200 µl of medium supplemented with 0.2% FBS and plated on the gels. The 24-well plate was filled with culture medium or serum-free medium. After 3–5 days, the embedded gel was sectioned without fixation and the invaded cells were analyzed in five different fields using fluorescence confocal microscopy (magnification, ×100; LSM710; Carl Zeiss AG). In these images, the distance of invaded cells was measured from five different positions and calculated using the ZEN blue edition program 1.1.2.0 version (Carl Zeiss AG). The distance in µm was calculated as previously described (31).
Three-dimensional (3D) Matrigel culture
The 3D cultures were observed as previously described (23). A total of 2×104 cells were suspended in a 400 µl medium supplemented with 2% Matrigel and seeded over a layer of 100% Matrigel in an 8-well culture slide (cat. no. 345108, Corning, Inc.). Cells were grown for 5 days and the medium was changed every 2 days. Colony formation was monitored every day and examined using a light microscope (magnification, ×100).
Measurement of Ras activation using ELISA
The cells were cultured to ~80% confluency in a 35 mm dish and were transfected for 24 h. The supernatants were removed and RAS activation was determined using a RAS activation ELISA kit (cat. no. 17-497; Millipore Sigma) according to the manufacturer's instructions. The level of activated RAS was compared with that of vector transfectant as a control. The results represent triplicated experiments.
Ras-GTP pull down assay
The cell lysates were prepared according to the manufacturer's recommendation (cat. no. BK008; Cytoskeleton, Inc.). The cells were cultured to ~80% confluency in a 60 mm dish and were transfected with the vectors at 37°C for 24 h. The cells were dissolved with the lysis buffer (cat. no. BK008; Cytoskeleton, Inc.) and centrifuged at 10,000 × g, 4°C for 1 min. The lysates (100 µg) were incubated with 30 µl of Raf-RBD beads at 4°C for 1 h and centrifuged at 5,000 × g, 4°C for 3 min. Then, the pellet was resuspended using 2X sample buffer (cat. no. BK008; Cytoskeleton, Inc.) and boiled at 95°C for 2 min. Samples were resolved using SDS-PAGE. Following resolution, procedures were the same as for immunoblotting. Antibodies for Ras (cat. no. 3965; Cell Signaling Technology, Inc.) K-Ras (cat. no. sc-30; Santa Cruz Biotechnology, Inc.), GST (cat. no. sc-138; Santa Cruz Biotechnology, Inc.) were used at 1:1,000 dilution.
Statistical analyses
Data were shown as the means ± standard deviation. Statistical analysis was assessed using the Student's t-test on the SigmaPlot software (Systat Software). Differences among three or more groups were estimated by one-way analysis of variance followed by Bonferroni multiple comparison tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Rab25 suppresses colon cancer cell invasion
In order to determine the role of Rab25 in colon cancer cell invasion, the cells were transfected with a control vector or Rab25 in colon cancer Caco-2 and HCT-116 cells. MTT analysis showed little difference in the viability between vector and Rab25 transfectants (Fig. S1). In addition, ectopic expression of Rab25 (Fig. S2) significantly attenuated the invasiveness (Fig. 1A) and the wound closure rates (Figs. 1B and S3) of these colon cancer cells. Rab25 markedly reduced HCT-116 cancer cell invasion (Fig. 1C). Furthermore, it was observed that Rab25 dramatically reduces the colony size of colon cancer cells on the 3D Matrigel system (Fig. 1D). To confirm the Rab25-induced inhibition of colon cancer cell invasion, the cells were transfected with Rab25 siRNA and it was noted that silencing of Rab25 (Fig. S4) significantly induced colon cancer cell invasion (Fig. 1E). Therefore, these data clearly indicated that Ra25 suppresses colon cancer cell invasion and migration.
Rab25 inhibits colon cancer cell EMT
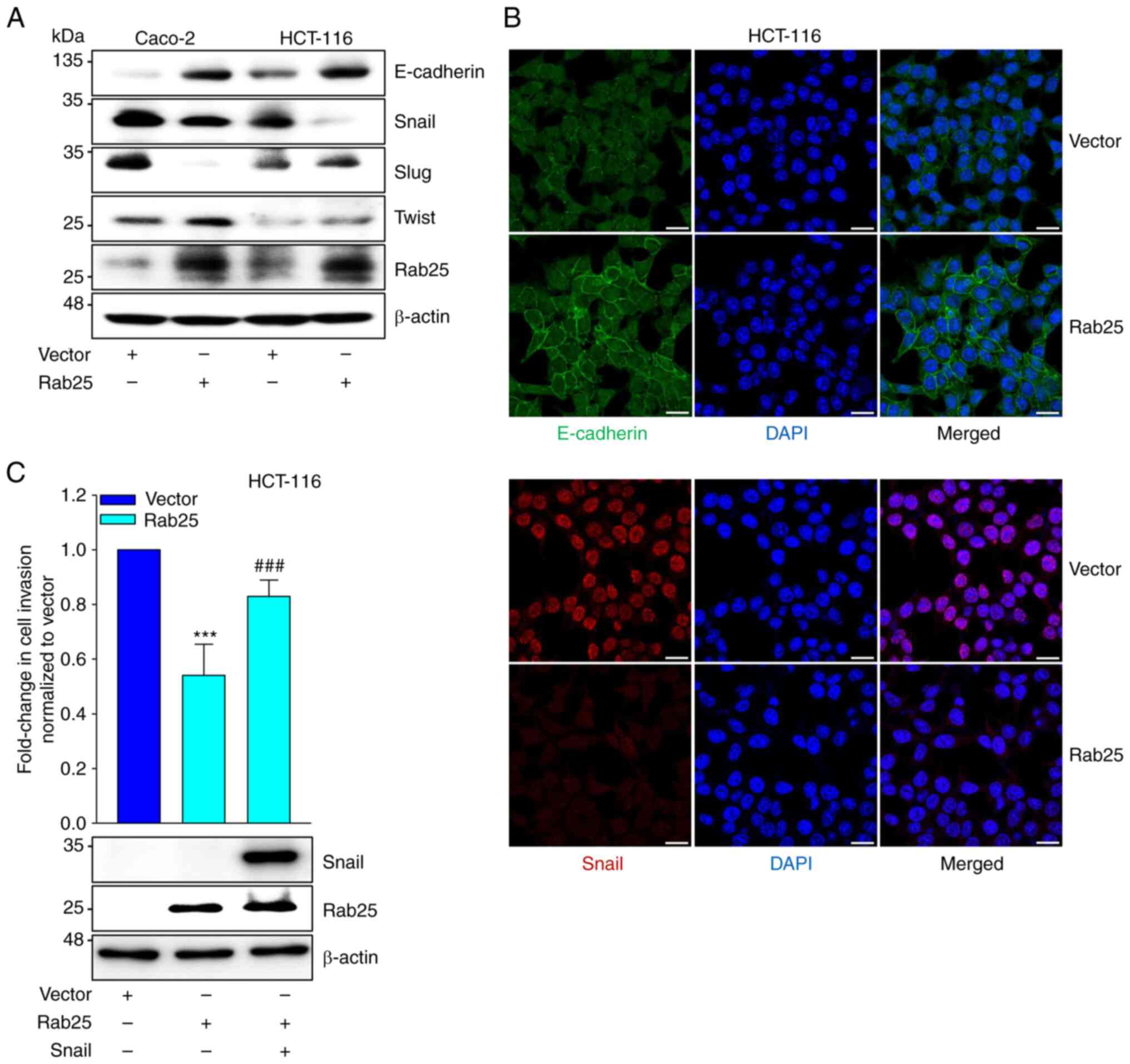
Given that an EMT transcription factor, Snail, is closely associated with Rab25-induced cancer cell invasiveness of various types of cancer (17), it was determined whether Rab25 regulated the expression of EMT factors. Immunoblotting data showed that Rab25 upregulated E-cadherin expression, while Snail expression was reduced by Rab25 (Fig. 2A). Consistently, immunofluorescence results demonstrated the upregulation of E-cadherin and downregulation of Snail expression by Rab25 in colon cancer cells (Fig. 2B). In contrast, silencing of Rab25 expression reduced and increased E-cadherin and Snail expression, respectively (Fig. S5). Furthermore, ectopic expression of Snail recovered colon cancer cell invasion repressed by Rab25 (Fig. 2C). Therefore, these data implied that Rab25 inhibited colon cancer cell EMT through the downregulation of Snail expression, leading to suppression of colon cancer cell invasion.
Rab25 inactivates EGFR
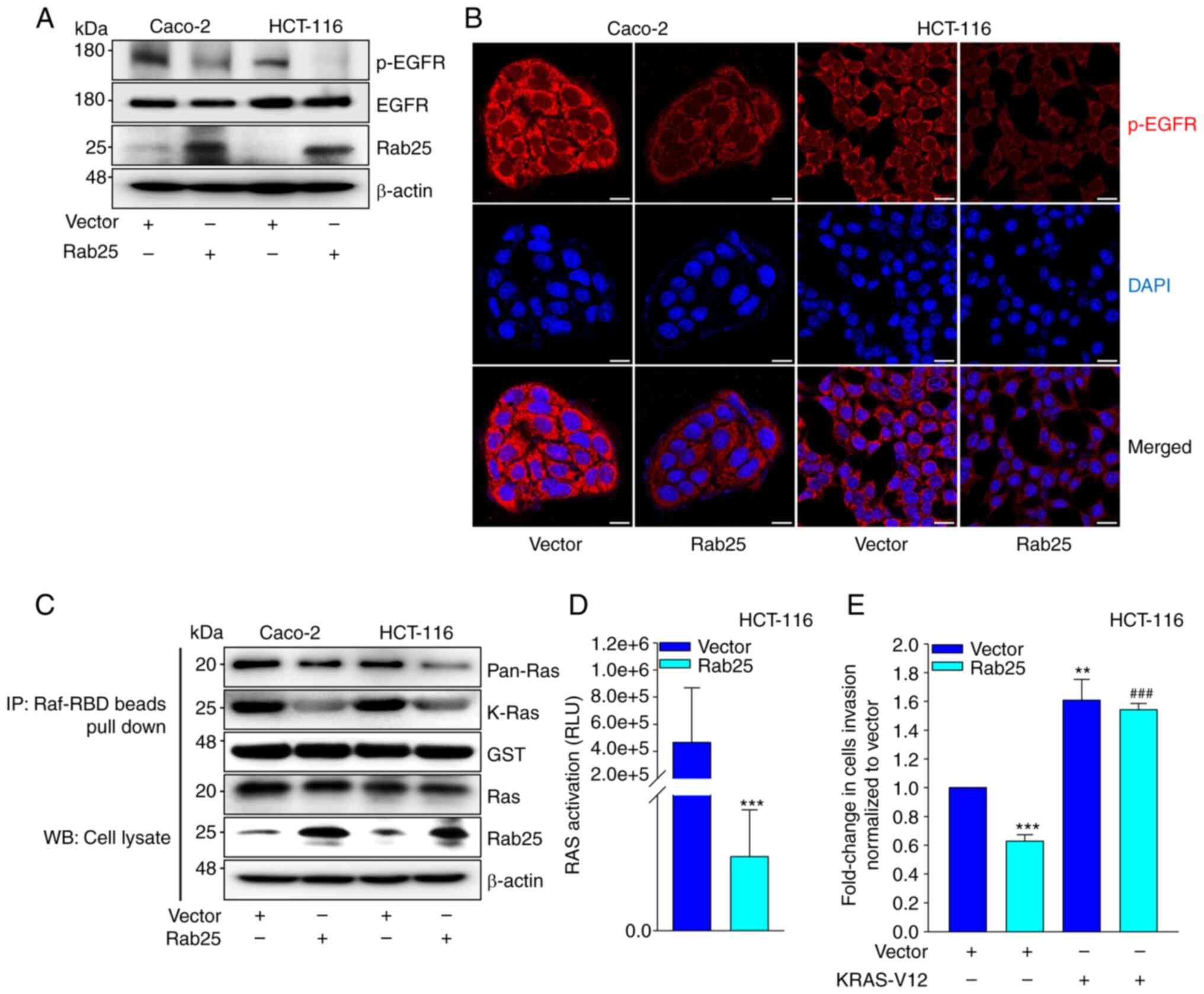
Previous data showed that EGFR is a critical upstream governor of Snail in Rab25-induced cancer cell invasion (17). In addition, EGFR has been closely associated with Rab25-induced cancer progression (17,32). Therefore, the involvement of EGFR in Rab25-induced suppression of colon cancer cell invasion was analyzed. Intriguingly, Rab25 profoundly reduced the levels of p-EGFR expression (Fig. 3A and B), suggesting that Rab25 inactivated EGFR and subsequent Snail expression for colon cancer cell invasion. Since one of downstream EGFR effectors is Ras, whether Rab25-induced suppression of EGFR activity influences Ras activity was determined. It was observed that Rab25 reduced Ras activity in colon cancer cells (Fig. 3C and D). In addition, ectopic expression of constitutively active KRAS (KRAS-V12) rescued Rab25-induced HCT-116 cell invasion (Fig. 3E), thus consolidating the involvement of Ras in Rab25-induced suppression of colon cancer cell invasion. Overall, these data indicated that Rab25 inhibits the EGFR/Ras/Snail signaling cascade to attenuate colon cancer cell invasion.
Rab25 induces claudin-7 expression
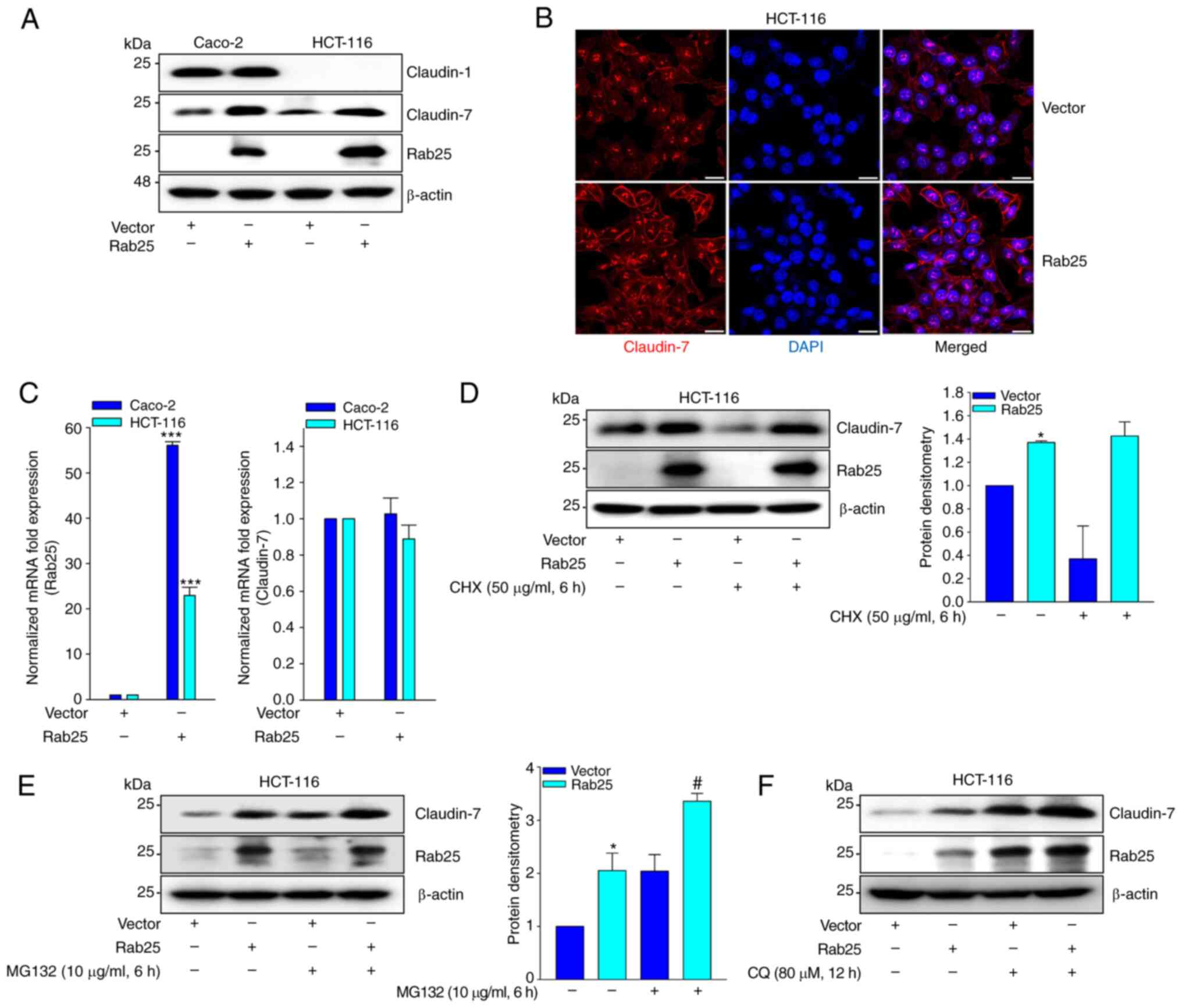
As claudin-7 induces Rab25 expression and suppresses colon cancer cell growth in a Rab25-dependent manner (16), the present study investigated whether Rab25 induces claudin-7 expression and consequently suppresses colon cancer cell invasion. Notably, Rab25 markedly increased claudin-7 expression (Fig. 4A and B). However, the present study did not observe Rab25 inducing claudin-7 transcript (Fig. 4C). Instead, Rab25 maintained the level of claudin-7 expression in the presence of CHX (Fig. 4D). Furthermore, treatment of the cells with pharmacological inhibitors of proteasome (MG-132; Fig. 4E) and lysosome (CQ; Fig. 4F) increased Rab25-induced claudin-7 expression. Collectively, these results suggested that Rab25 induces claudin-7 expression by stabilizing its protein.
Claudin-7 mediates Rab25-induced suppression of colon cancer cell invasion
Next, the role of claudin-7 in Rab25-induced suppression of colon cancer cell invasion was explored. Ectopic expression of claudin-7 reduced the expression of p-EGFR and Snail (Fig. 5A). In addition, claudin-7 upregulated E-cadherin expression (Fig. S6) and attenuated colon cancer cell invasion, which was reversed by enforced Snail expression (Fig. 5B). Consistently, silencing of claudin-7 expression increased colon cancer cell invasion (Fig. 5C) and migration (Fig. 5D). Furthermore, claudin-7 siRNA recovered Rab25-induced EGFR inactivation and suppression of Snail expression (Fig. 5E). In addition, it was observed that claudin-7 significantly reduced Ras activity in colon cancer cells (Fig. 5F) and that ectopic expression of constitutively active KRAS (KRAS-V12) rescued claudin-7-induced suppression of HCT-116 cell invasion (Fig. 5G). Therefore, these data implied that claudin-7 mediated Rab25-induced attenuation of colon cancer cell invasion.
Discussion
Accumulating evidence suggests the context-dependent role of Rab25 in cancer cell progression. We previously showed that Rab25 aggravates cancer cell invasion through the β1 integrin/EGFR/Snail signaling axis (17). the present study demonstrated that Rab25 suppressed colon cancer cell invasion by upregulating claudin-7 expression. Rab25 and claudin-7 both inhibited EGFR activation and EMT. In addition, these proteins attenuated colon cancer cell invasion. Importantly, the data showed that Rab25 induced claudin-7 expression via protein stabilization, uncovering the critical role of these proteins in regulating colon cancer cell invasion.
Rab25 has been demonstrated to induce cancer cell invasion by upregulating β1 integrin and subsequent activation of the EGFR/VEGF/Snail signaling axis in ovarian, stomach and estrogen receptor-positive breast cancer (17). Although Rab25 attenuates cancer invasiveness through upregulating β1 integrin expression (8,33–35), the detailed underlying mechanism remains to be elucidated. The present study identified the critical role of claudin-7 in Rab25-induced suppression of colon cancer cell invasion. First, Rab25 induced claudin-7 expression through protein stabilization. Second, Rab25 inactivated EGFR in a claudin-7-dependent manner; the silencing of claudin-7 rescued Rab25-induced EGFR inactivation. In addition, Rab25 downregulated Snail expression, which is important for colon cancer cell EMT and invasiveness. Furthermore, claudin-7 alone reduced Snail expression and, consequently, attenuated colon cancer cell invasion.
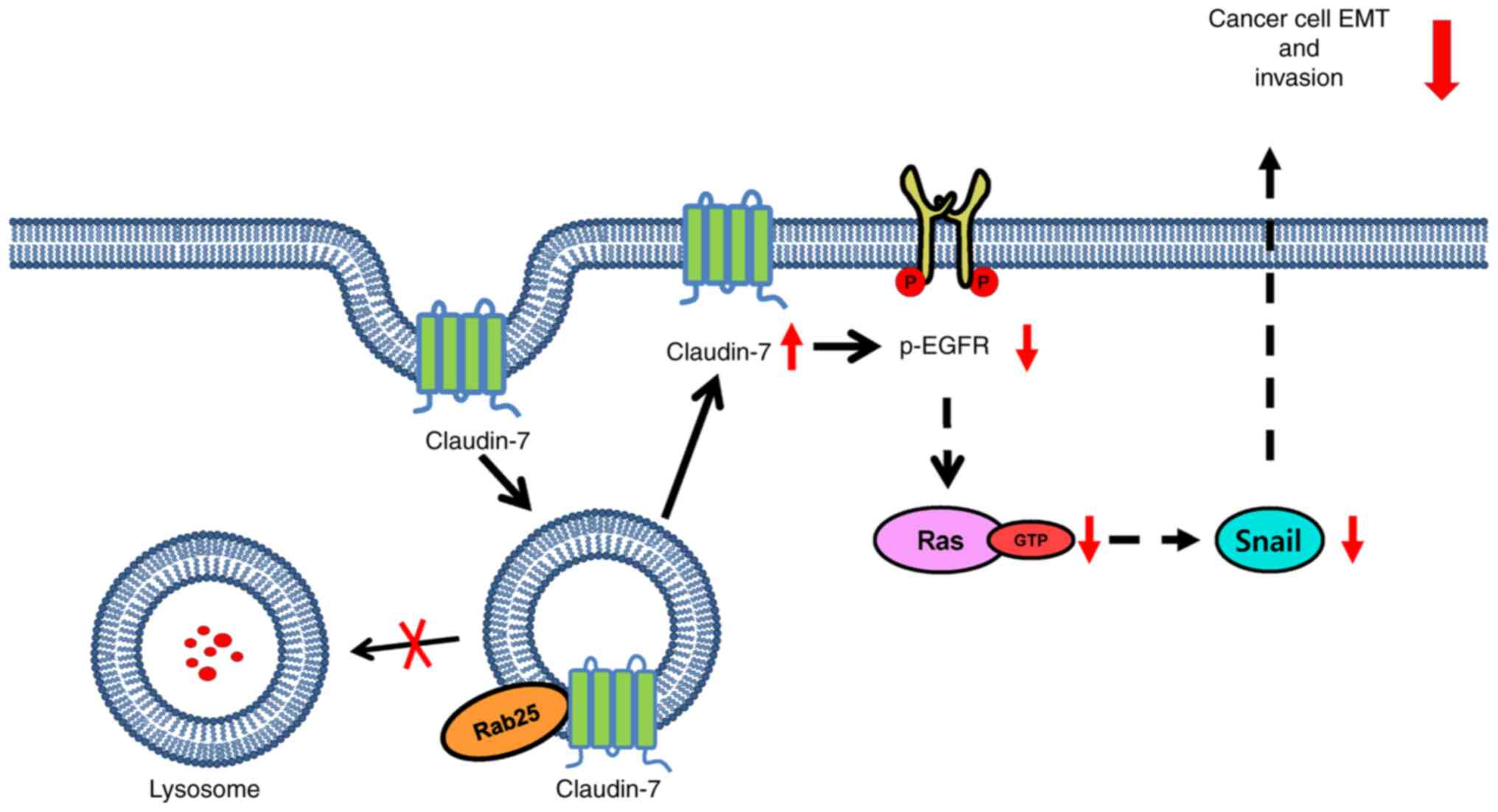
Claudin-7 is a member of the family of tight junction proteins and is implicated in the suppression of proliferation and invasion of various types of cancer including colon cancer (15,16,36). Similarly, claudin-7 was shown to induce Rab25 expression to suppress colon cancer cell tumorigenesis and invasion (16). The present study observed that Rab25 in turn increased claudin-7 protein expression. Although claudin-7 was shown to induce Rab25 expression through transcriptional activation (16), the data demonstrated that Rab25 increased claudin-7 protein expression without any effect on mRNA expression. At present, it is not known how Rab25 protects claudin-7 from proteasomal degradation. Since the present study showed that CQ maintained claudin-7 protein levels (Fig. 4F), Rab25 might recycle claudin-7 in the late endosome to the plasma membrane, thus preventing its lysosomal degradation, which is similar to what happens to β1 integrin. Notably, a previous study suggests that claudin-7 forms a protein complex with β1 integrin in lung cancer cells (15), which reinforces the hypothesis that Rab25 protects both β1 integrin and claudin-7 from lysosomal degradation through recycling endosomes to the plasma membrane and subsequent inactivation of the EGFR/Snail axis for colon cancer cell invasion. A further detailed mechanistic study is currently underway. Fig. 6 illustrates the results schematically in which Rab25 salvages claudin-7 from lysosomal degradation and thereby inhibits the EGFR/Ras/Snail signaling axis to attenuate colon cancer cell invasion. Collectively, our present study demonstrated a novel role of claudin-7 in Rab25-induced suppression of colon cancer cell invasion by inactivating the EGFR/Snail axis, thus providing crucial biomarkers and therapeutic potential for colon cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grant from the Basic Science Research Program of the National Research Foundation of Korea grant, funded by the Ministry of Education, Science and Technology (grant no. NRF-2022R1A2C1004019).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL and GP conceived and designed the present study. Data curation was performed by SC and BJ. Formal analysis was performed by SC. Investigation was by SC, BJ and SY. Methodology was by SC and BJ. Supervision was by HL and GP. Writing of original draft was by HL. Writing and editing was by HL and SC. All authors discussed the results and commented on the manuscript. SC and BJ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent of participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
Rab25 |
Ras-related protein 25 |
|
EGFR |
epidermal growth factor receptor |
|
EMT |
epithelial to mesenchymal transition |
|
VEGF |
vascular endothelial growth factor |
|
MTT |
(3,4,5-dimethylathiazol-2yl)-5-diphenyl-tetrazolium bromide |
|
CHX |
cycloheximide |
|
CQ |
chloroquine |
References
|
Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al: The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 10:1251–1256. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng JM, Ding M, Aribi A, Shah P and Rao K: Loss of RAB25 expression in breast cancer. Int J Cancer. 118:2957–2964. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, et al: Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 13:496–510. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Wang S, Hu C, Wu F and He S: Rab25 GTPase: Functional roles in cancer. Oncotarget. 8:64591–64599. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Calhoun BC and Goldenring JR: Rab proteins in gastric parietal cells: Evidence for the membrane recycling hypothesis. Yale J Biol Med. 69:1–8. 1996.PubMed/NCBI | |
|
Cao C, Lu C, Xu J, Zhang J and Li M: Expression of Rab25 correlates with the invasion and metastasis of gastric cancer. Chin J Cancer Res. 25:192–199. 2013.PubMed/NCBI | |
|
Zhang J, Wei J, Lu J, Tong Z, Liao B, Yu B, Zheng F, Huang X, Chen Z, Fang Y, et al: Overexpression of Rab25 contributes to metastasis of bladder cancer through induction of epithelial-mesenchymal transition and activation of Akt/GSK-3β/Snail signaling. Carcinogenesis. 34:2401–2408. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Nam KT, Lee HJ, Smith JJ, Lapierre LA, Kamath VP, Chen X, Aronow BJ, Yeatman TJ, Bhartur SG, Calhoun BC, et al: Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. J Clin Invest. 120:840–849. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Cho KH and Lee HY: Rab25 and RCP in cancer progression. Arch Pharm Res. 42:101–112. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, Cao J and Chen YH: Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology. 142:305–315. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Wang K, Xu C, Li W and Ding L: Emerging clinical significance of claudin-7 in colorectal cancer: A review. Cancer Manag Res. 10:3741–3752. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Nakayama F, Semba S, Usami Y, Chiba H, Sawada N and Yokozaki H: Hypermethylation-modulated downregulation of claudin-7 expression promotes the progression of colorectal carcinoma. Pathobiology. 75:177–185. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Oshima T, Kunisaki C, Yoshihara K, Yamada R, Yamamoto N, Sato T, Makino H, Yamagishi S, Nagano Y, Fujii S, et al: Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol Rep. 19:953–959. 2008.PubMed/NCBI | |
|
Wang K, Li T, Xu C, Ding Y, Li W and Ding L: Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem Biophys Res Commun. 508:797–804. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Lu Z, Kim DH, Fan J, Lu Q, Verbanac K, Ding L, Renegar R and Chen YH: A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer. 14:1202015. View Article : Google Scholar : PubMed/NCBI | |
|
Bhat AA, Pope JL, Smith JJ, Ahmad R, Chen X, Washington MK, Beauchamp RD, Singh AB and Dhawan P: Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene. 34:4570–4580. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Jeong BY, Cho KH, Jeong KJ, Park YY, Kim JM, Rha SY, Park CG, Mills GB, Cheong JH and Lee HY: Rab25 augments cancer cell invasiveness through a β1 integrin/EGFR/VEGF-A/Snail signaling axis and expression of fascin. Exp Mol Med. 50:e4352018. View Article : Google Scholar : PubMed/NCBI | |
|
Mitra S, Federico L, Zhao W, Dennison J, Sarkar TR, Zhang F, Takiar V, Cheng KW, Mani S, Lee JS, et al: Rab25 acts as an oncogene in luminal B breast cancer and is causally associated with Snail driven EMT. Oncotarget. 7:40252–40265. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Jeong BY, Park SR, Cho S, Yu SL, Lee HY, Park CG, Kang J, Jung DY, Park MH, Hwang WM, et al: TGF-β-mediated NADPH oxidase 4-dependent oxidative stress promotes colistin-induced acute kidney injury. J Antimicrob Chemother. 73:962–972. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Cho KH, Jeong BY, Park CG and Lee HY: The YB-1/EZH2/amphiregulin signaling axis mediates LPA-induced breast cancer cell invasion. Arch Pharm Res. 42:519–530. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Jeong BY, Cho KH, Jeong KJ, Cho SJ, Won M, Kim SH, Cho NH, Hur GM, Yoon SH, Park HW, et al: Lysophosphatidic acid-induced amphiregulin secretion by cancer-associated fibroblasts augments cancer cell invasion. Cancer Lett. 551:2159462022. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Hwang MH, Cho KH, Jeong KJ, Park YY, Kim JM, Yu SL, Park CG, Mills GB and Lee HY: RCP induces slug expression and cancer cell invasion by stabilizing β1 integrin. Oncogene. 36:1102–1111. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Choe SR, Kim YN, Park CG, Cho KH, Cho DY and Lee HY: RCP induces FAK phosphorylation and ovarian cancer cell invasion with inhibition by curcumin. Exp Mol Med. 50:1–10. 2018. View Article : Google Scholar | |
|
Cho SJ, Jeong BY, Song YS, Park CG, Cho DY and Lee HY: STAT3 mediates RCP-induced cancer cell invasion through the NF-κB/Slug/MT1-MMP signaling cascade. Arch Pharm Res. 45:460–474. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Park SY, Jeong KJ, Panupinthu N, Yu S, Lee J, Han JW, Kim JM, Lee JS, Kang J, Park CG, et al: Lysophosphatidic acid augments human hepatocellular carcinoma cell invasion through LPA1 receptor and MMP-9 expression. Oncogene. 30:1351–1359. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Kim YH, Choi YW, Lee J, Soh EY, Kim JH and Park TJ: Senescent tumor cells lead the collective invasion in thyroid cancer. Nat Commun. 8:152082017. View Article : Google Scholar : PubMed/NCBI | |
|
Satoyoshi R, Kuriyama S, Aiba N, Yashiro M and Tanaka M: Asporin activates coordinated invasion of scirrhous gastric cancer and cancer-associated fibroblasts. Oncogene. 34:650–660. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Tanaka M, Kuriyama S, Itoh G, Kohyama A, Iwabuchi Y, Shibata H, Yashiro M and Aiba N: Identification of anti-cancer chemical compounds using xenopus embryos. Cancer Sci. 107:803–811. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Kim JY, Cho KH, Jeong BY, Park CG and Lee HY: Zeb1 for RCP-induced oral cancer cell invasion and its suppression by resveratrol. Exp Mol Med. 52:1152–1163. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Vehlow A, Klapproth E, Storch K, Dickreuter E, Seifert M, Dietrich A, Bütof R, Temme A and Cordes N: Adhesion- and stress-related adaptation of glioma radiochemoresistance is circumvented by β1 integrin/JNK co-targeting. Oncotarget. 8:49224–49237. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang L, Xie B, Qiu Y, Jing D, Zhang J, Duan Y, Li Z, Fan M, He J, Qiu Y, et al: Rab25-mediated EGFR recycling causes tumor acquired radioresistance. iScience. 23:1009972020. View Article : Google Scholar : PubMed/NCBI | |
|
Goldenring JR and Nam KT: Rab25 as a tumour suppressor in colon carcinogenesis. Br J Cancer. 104:33–36. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Krishnan M, Lapierre LA, Knowles BC and Goldenring JR: Rab25 regulates integrin expression in polarized colonic epithelial cells. Mol Biol Cell. 24:818–831. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Hong KS, Jeon EY, Chung SS, Kim KH and Lee RA: Epidermal growth factor-mediated Rab25 pathway regulates integrin β1 trafficking in colon cancer. Cancer Cell Int. 18:322018. View Article : Google Scholar : PubMed/NCBI | |
|
Kim DH, Lu Q and Chen YH: Claudin-7 modulates cell-matrix adhesion that controls cell migration, invasion and attachment of human HCC827 lung cancer cells. Oncol Lett. 17:2890–2896. 2019.PubMed/NCBI |