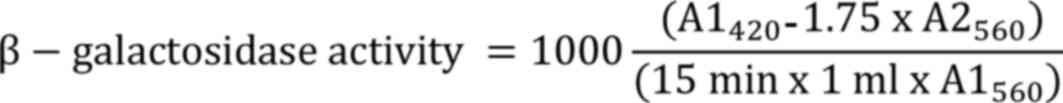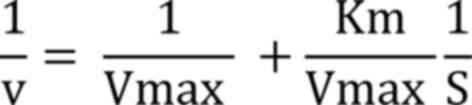Kinetic and modeling analyses of lactose‑hydrolyzing β‑galactosidase from Lactiplantibacillus plantarum GV54
- Authors:
- Published online on: January 31, 2023 https://doi.org/10.3892/wasj.2023.188
- Article Number: 11
-
Copyright: © Vasudha et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Lactic acid bacteria (LAB), which are employed as starters in the manufacturing of dairy products, are critical variables in the fermentation and preservation of fermentative foods, improving the texture and flavor of food products (1). Fermented foods containing LAB are traditionally used every day as food in several parts of India. Curd, prepared by the fermentation of milk with an inoculum of previously prepared curd, is used in the majority of households in India, where it constitutes an integral part of the daily diet. The LAB that ferment the milk are likely to differ slightly in each household, as there is no standardized starter culture used to prepare curd. Although curd is believed to have probiotic properties, the documentation of this is limited of this in the literature (2). The present study was undertaken to evaluate the LAB from homemade curd in southern India for its probiotic properties. Rural symbolizes the ethnicity of food culture, having indigenous microbial culture affording to improve the general health of consumers. The intention of collecting samples from such a region was to obtain the bacteria which are prototrophs. Although there are not many differences as regards homes or locations, the diversity of LAB perhaps yields novel enzyme production.
β-galactosidase (EC 3.2.1.23) is the most essential enzyme in the dairy sector for the production of low-lactose foods to address lactose intolerance. It is produced commercially from microorganisms, such as bacteria, yeast and fungus. Bacterial enzymes are preferred over synthetic ones due to their high activity and stability (3,4). β-galactosidase is employed in the food industry to hydrolyze lactose into glucose and galactose, preventing lactose crystallization and enhancing the sweetness and milk product solubility. In addition, β-galactosidase has two distinct enzymatic functions: It breaks down lactose and also cleaves cellobiose, cellotriose, and cellulose to a lesser extent (5). The majority of chemically synthesized β-galactosidase is not approved for use in food, as it is expensive and less efficient for industrial usage. Therefore, there is an urgent need to address the gap by selecting microorganisms which are safe for consumption and capable of producing β-galactosidase. It has been observed that, for functional food applications, LAB and Bifidobacterium, are safe organisms and the most desirable sources of β-galactosidase (6).
β-galactosidase has various applications in the fields of medicine and industry. A biosensor can be developed using the β-galactosidase enzyme to determine the lactose in milk, and it is also used in the diagnosis and treatment of lactose intolerance. The inability to digest lactose in the small intestine due to a low β-galactosidase activity known as lactose intolerance, is associated with clinical symptoms such as nausea, bloating, flatulence and/or stomach discomfort after consuming lactose or dietary components containing lactose; this may affect everyday activities and the quality of life of individuals with this condition (5,7). The encapsulation of the enzyme may be the strategy for the production of lactose-hydrolyzed milk (8,9). The first stage in isolating the intracellular enzyme, β-galactosidase, from LAB, is to interrupt the cell by various mechanical and chemical methods (10). In addition, the enzyme extracted from Aspergillus niger (A. niger) is useful in the commercial production of an array of sugars, such as glucose, galactose, heteropolysaccharides, galacto-oligosaccharide. The β-galactosidase enzyme from A. niger has undergone analyses regarding comparative modeling, structural annotation, domain identification and structural comparison. Additionally, the catalytic sites, Glu200 and Glu298, with β-D-galactose were docked independently (11-14).
The main objective of the present study was to produce β-galactosidase from LAB and a selection of Lactiplantibacillus isolates producing potential β-galactosidase. By biochemical analysis [with 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-Gal) and O-nitrophenyl-β-D-galactopyranoside (ONPG) as substrates] and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the enzyme was characterized. For enzyme optimization, physical conditions (pH and temperature), the effects of metal ions and kinetics with various ONPG substrate concentrations were examined. In addition, the construction of a 3D structure, structural annotation and structural comparison of the β-galactosidase enzyme was determined.
Materials and methods
Isolation and screening of LAB. A total of 16 homemade curd samples were collected from rural areas in Davangere, Karnataka, India. The de Man, Rogosa and Sharpe (MRS) agar medium (Himedia Laboratories Pvt., Ltd.) was used for the isolation of LAB and incubated at 37˚C for 24 to 48 h. The isolated colonies were purified by several successive streaks on MRS medium and the selected bacterial isolates were preserved at 4˚C on MRS slants until further use. The ability of the bacterial isolate to produce β-galactosidase was examined using the X-Gal (Himedia Laboratories Pvt., Ltd.) substrate supplemented with MRS medium and isopropyl β-D-1-thiogalactosidase (IPTG) (Himedia Laboratories Pvt., Ltd.) as an inducer for β-galactosidase. The production of blue-colored colonies indicated the production of β-galactosidase and were selected for further analyses. In another study (15), the authors isolated and screened 27 β-galactosidase-producing isolates, further subjected to morphological and physiological characterization. Furthermore, the eight most potential probiotic β-galactosidase-producing LAB were identified using 16S rRNA gene sequencing. All the eight isolates belong to the phylum Firmicutes, exhibiting the highest similarity with Lactobacillus spp. Amongst these, four LAB producing high concentrations of β-galactosidase were selected for use in the present study.
Production of β-galactosidase enzyme. For the production of β-galactosidase, MRS culture medium was supplemented with 4% lactose. The selected bacterial culture was inoculated in 100 ml MRS medium and incubated at 37˚C for 48 h (200 x g). The bacterial cells were further harvested at 12,000 x g for 5 min at 4˚C and suspended in phosphate buffer (pH 6.8) to measure the activity of β-galactosidase, as previously described (16).
Optimization of β-galactosidase using different carbon and nitrogen sources. The effects of media composition on β-galactosidase production were determined by using different carbon sources, such as arabinose, fructose, glucose, galactose, lactose, sucrose, mannose and xylose. Additionally, ammonium chloride, ammonium nitrate, ammonium sulfate, peptone, casein, urea, yeast extract, beef extract, sodium and potassium nitrate (Himedia Laboratories Pvt., Ltd.) were used as nitrogen sources for the optimization of production media.
Extraction and partial purification of intracellular β-galactosidase. The extraction of enzyme was performed using the cell permeabilized method with the toluene/acetone solvent system. A total of 10 ml bacterial culture was treated with 0.1 ml toluene/acetone (1:9 v/v) (Himedia Laboratories Pvt., Ltd.) and vortexed for 7 min, as previously described (15). The cell-free supernatant was precipitated with ammonium sulfate (Himedia Laboratories Pvt., Ltd.) using a magnetic stirrer until it reached a saturation level of 70% (w/v). That precipitate was then centrifuged at 12,000 x g, for 10 min at 4˚C using 0.1 M phosphate buffer (Na2HPO4 x 7H2O/40 mM NaH2PO4 buffer pH 7.0) and the obtained precipitate was dialyzed overnight against a phosphate buffer (Himedia Laboratories Pvt., Ltd.).
SDS-PAGE. A 12% SDS-PAGE was used as the resolving gel for the enzyme analysis. A pre-stained protein ladder (MBT092-10LN) was purchased from HiMedia Laboratories Pvt., Ltd. The gel was run for 4 h at 110 V and the enzyme bands were visualized by staining with 0.25% Coomassie brilliant blue G-250 at 28˚C for 1 h (Himedia Laboratories Pvt., Ltd.) and de-stained with a mixture of 10% acetic acid and 40% methanol (Himedia aboratories Pvt., Ltd.).
β-Galactosidase assay. For the β-galactosidase assay, the reaction mixture consisted of 200 µl ONPG and 100 µl of the extracted enzyme with 900 µl of 60 mM phosphate buffer (Himedia Laboratories Pvt., Ltd.) (pH 7). Subsequently, 0.5 ml of 1 M sodium carbonate (NaCO3) was added to the reaction mixture and incubated for 15 min at 37˚C to terminate the reaction, which was then measured at 420 and 560 nm using a NanoDrop 2000C UV-Spectrophotometer (Thermo Fisher Scientific, Inc.) and β-galactosidase activity was represented in Miller units as follows:
where, A1560 was the absorbance value of the reaction mixture prior to incubation, and A1420 and A2560 were the absorbance values of the reaction mixture following incubation.
Characterization of β-galactosidase enzyme
Effects of pH and temperature. The effects of pH and temperature on intracellular β-galactosidase were determined using ONPG as a substrate. The reaction mixture containing partially purified β-galactosidase with 15 mM ONPG and various pH levels (pH 5.0 to 9.0) was adjusted (pH 5 0.1 M sodium acetate buffer, pH 6-7 0.1 M sodium phosphate buffer, and pH 8-9 0.1 M Tris HCl buffer), and further incubated at 37˚C for 15 min. Similarly, β-galactosidase was incubated for 15 min at various temperatures 30, 35, 37, 40, 45 and 50˚C, set up to determine the maximum enzyme activity.
Effects of various metal ions. To examine the effects of various cations on β-galactosidase activity (release of O-nitrophenol from ONPG), the enzyme samples were assayed with ONPG in the presence 1.0 mM of various metal ions [sodium chloride (NaCl), potassium chloride (KCl), silver nitrate (AgNO3), magnesium sulfate (MgSO4), calcium chloride (CaCl2), copper sulfate (CuSO4), manganese(II) chloride (MnCl2) and ferrous sulfate (FeSO4) compound form] and the enzyme activity was determined as mentioned above.
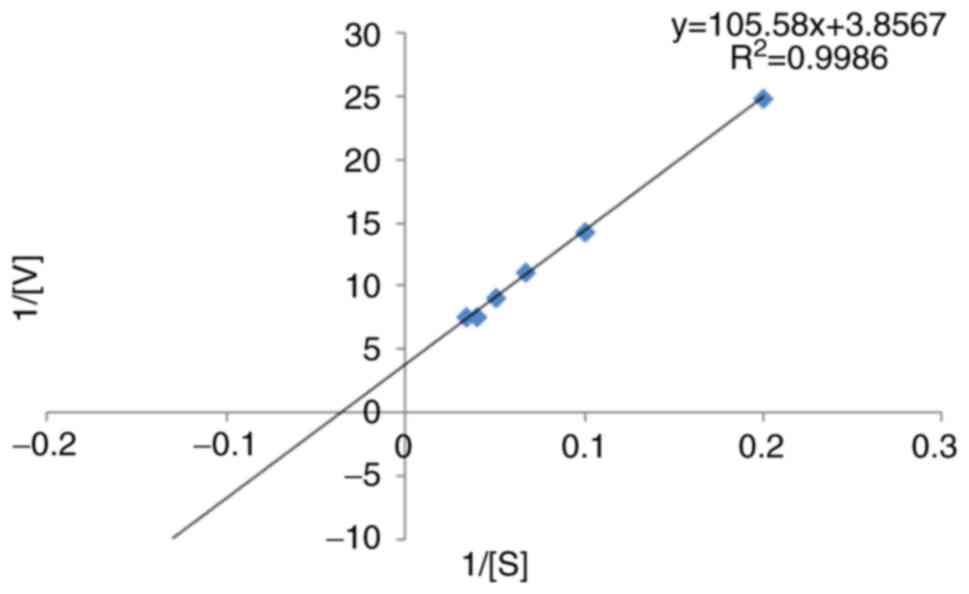
Kinetics analysis. Enzyme kinetics were determined using ONPG as a substrate range between 1-22 mM at pH 7.0, and Lineweaver-Burk plot (1/V against 1/S) was used to measure the maximum velocity (Vmax) and kinetic constant (Km) using following equation:
3D modeling and structural annotation of β-galactosidase
Sequence retrieval. The National Centre for Biotechnology Information's GenPept database was used to identify the amino acid sequence of the β-galactosidase enzyme from Lactiplantibacillus plantarum in FASTA format (L. plantarum; http://www.ncbi.nlm.nih.gov/protein/; https://www.ncbi.nlm.nih.gov/genbank/fastaformat/).
Comparative modeling and structural annotation. An important and promising field of study is the prediction of protein structure via comparative modeling (13), using the 3D jigsaw server for the comparative modeling of the target protein, and the SAS-sequence annotated by the structure server (http://www.ebi.ac.uk/thornton-srv/databases/sas/), which was used to characterize the structural annotation. The predicted structure was visualized using the PyMol program. (https://pymol.org/2/).
ProFunc, a database of protein functions from the Thornton Structure Server (http://www.ebi.ac.uk/thornton-srv/databases/ProFunc/), and PDBsum, a visual representation of 3D protein structures from the Protein Data Bank server (http://www.ebi.ac.uk/pdbsum/), are both accessible through the European Bioinformatics Institute was also used.
Structural comparison. The similarities and contrasts of related homologous-3D structures are frequently emphasized using protein structural comparisons. Although they share a common ancestor protein, homologous proteins, although multiplied, develop along different pathways, and are throughout time (13,14). In order to identify comparable structures in other organisms, a structural comparison of the modeled structure was carried out using the Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/) in the PDB database (14).
Statistical analysis. All results are reported as the mean ± standard error of mean and GraphPad Prism 9 software (GraphPad Software; Dotmatics) was used to compare data using one-way ANOVA with post hoc Tukey's multiple comparisons test. 3D structure analyses were performed using EMBL-EBI database software.
Results
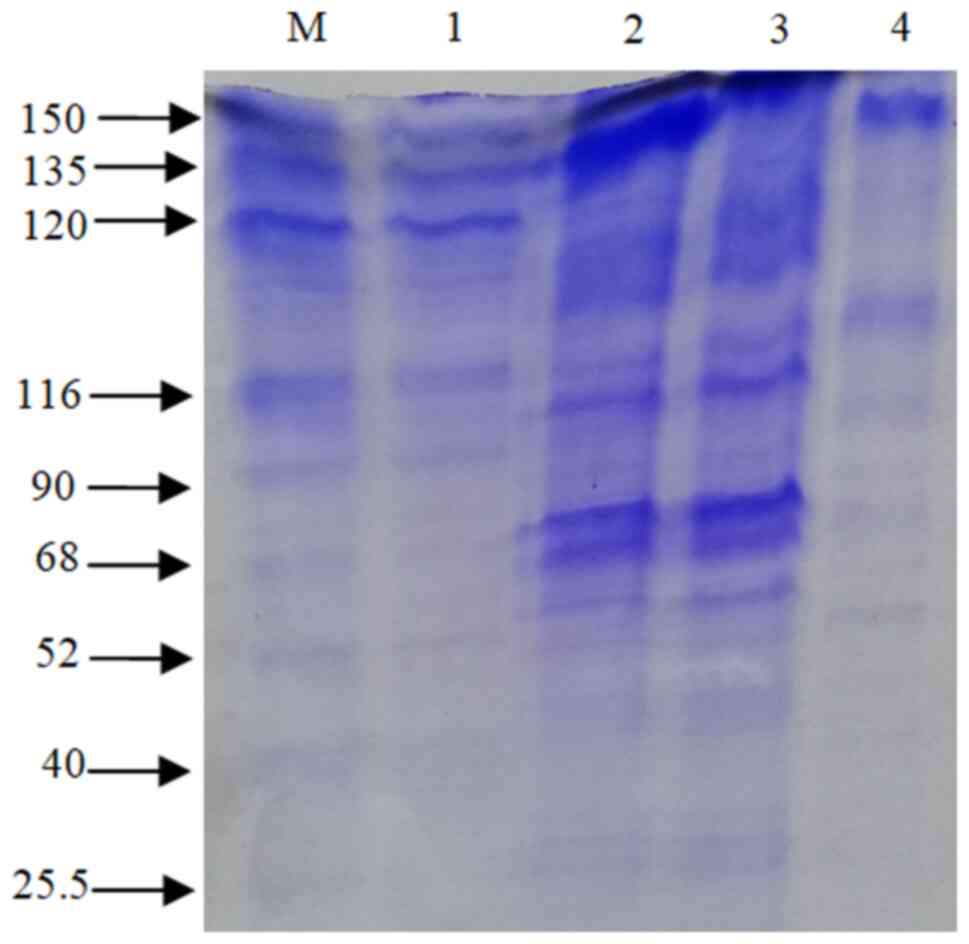
A total of 190 LAB were isolated from 16 different homemade curd samples and screened for β-galactosidase production using X-Gal substrate. A total of 27 isolates exhibited blue coloration on MRS agar medium supplemented with the X-Gal substrate. The data presented in Table I revealed that L. plantarum GV54 (56.32±0.6 Miller units; P<0.05) and L. plantarum GV64 (48.35±1.63 Miller units; P<0.05) significantly increased β-galactosidase production by utilizing lactose as a carbon source compared with other different carbon sources tested. L. plantarum GV54 exhibited maximum enzyme production by utilizing ammonium sulfate and beef extract (31.12±0.05 and 28.45±0.95 Miller units; P<0.05 respectively) as two nitrogen sources compared with other different nitrogen sources tested (Table II). β-galactosidase was extracted using the cell permeabilization method with toluene/acetone solvents. SDS-PAGE (12%) was used for band separation and the molecular weight was determined (~116 kDa) (Fig. 1). The β-galactosidase activity of L. plantarum GV54 was comparably high in both qualitative and quantitative assays. The maximum enzyme activity (162.96±0.36 Miller units; P<0.05) was exhibited by the L. plantarum GV54 isolate compared to the other isolates tested (Table III). As illustrated in Figs. 2 and 3, β-galactosidase activity was high at pH 7.0 and 37˚C. The potential of various cations to either stimulate or inhibit the enzyme activity was investigated. As per the data presented in Table IV, the addition of Mg2+, Mn2+, and Fe2+ had an indirect effect on the relative enzymatic activity, and Mn2+ was shown to lead to the maximum relative enzymatic activity. However, the addition of Ag+ decreased the enzyme activity. However, these results need to be validated in the future using in vivo animal model studies or human clinical trials in collaboration with health departments.
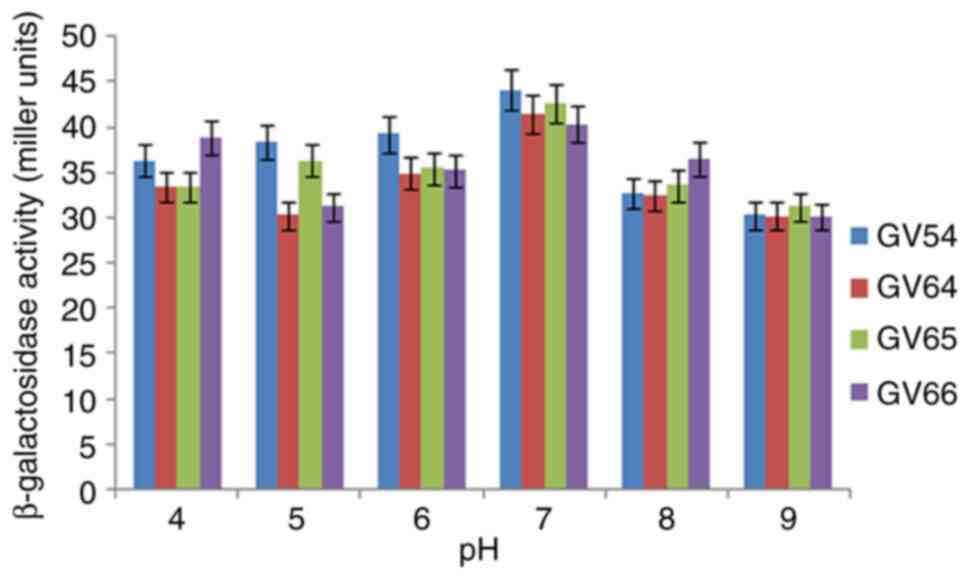 | Figure 2Effect of various pH levels on β-galactosidase activity extracted from lactic acid bacteria. |
Table IEffect of carbon sources on intracellular β-galactosidase production by Lactiplantibacillus isolates. |
Table IIEffect of nitrogen sources on intracellular β-galactosidase production by Lactiplantibacillus isolates |
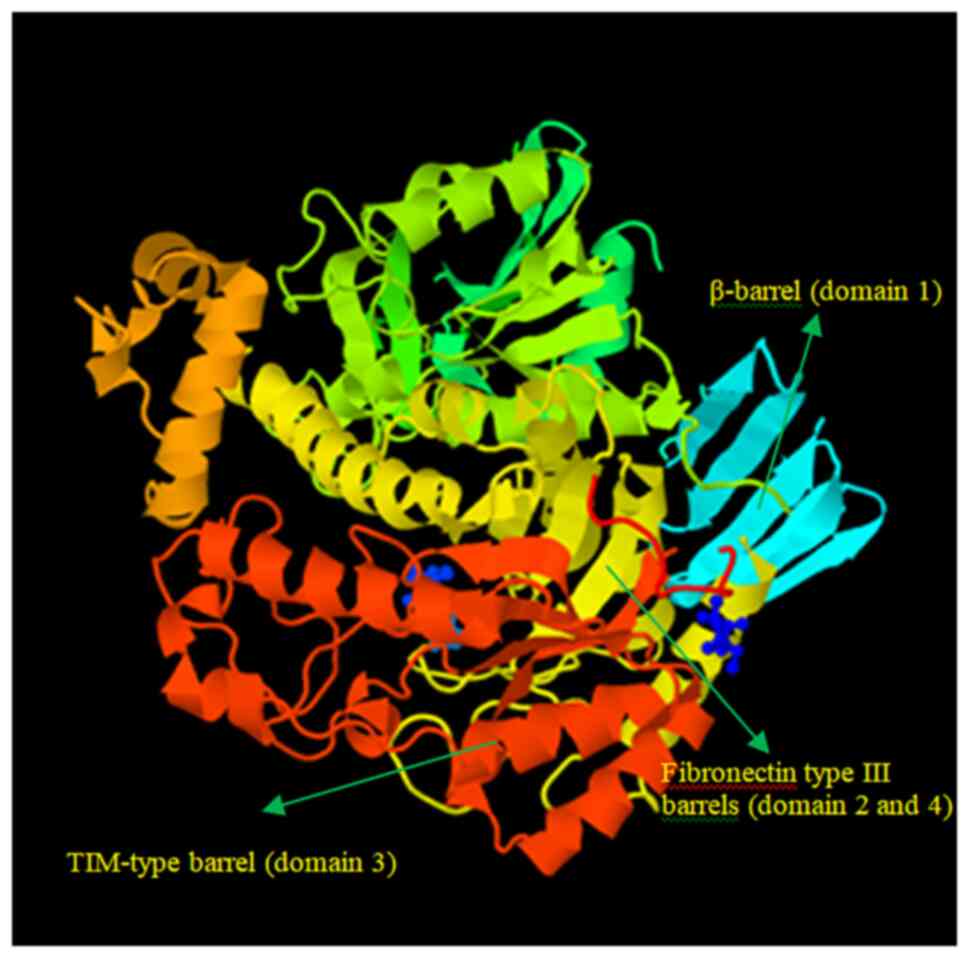
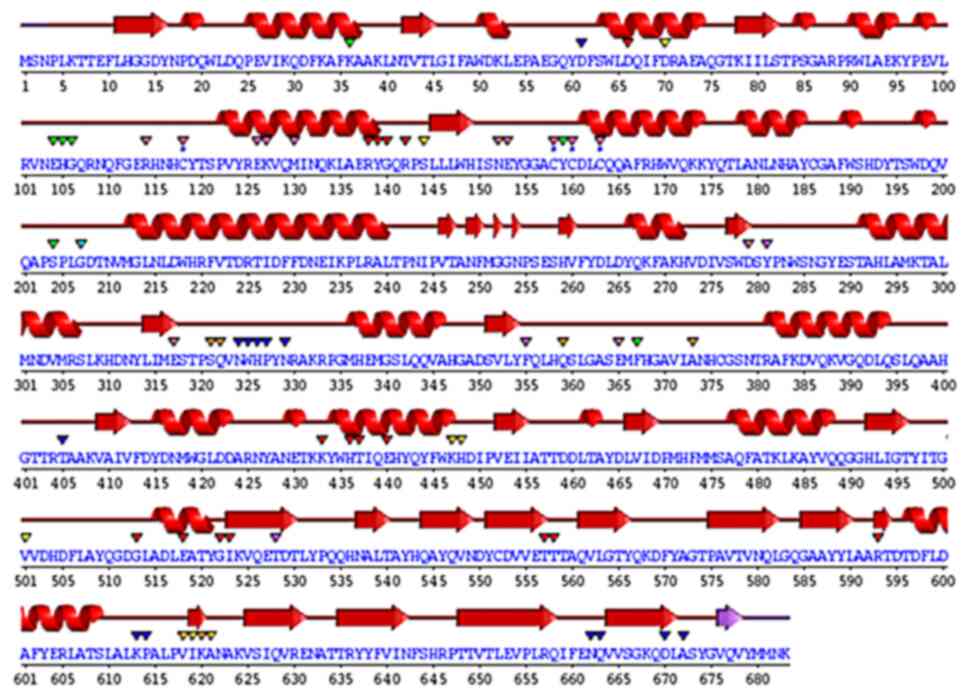
The kinetic parameters of β-galactosidase with the ONPG substrate were determined using the Lineweaver-Burk plot. The Km was found to be 27.3757 mM, and its Vmax was 0.2592 U/min (Fig. 4). Using the FASTA format, the amino acid sequence of β-galactosidase (accession no. KZD98639.1) was retrieved, which was isolated from L. plantarum. By employing a comparison modeling method, the 3D jigsaw server produced the 3D structure of the target protein. The PyMOL application was used to view the anticipated structure (Fig. 5). The wiring diagram presented in Fig. 6 was produced by the SAS server. The crystal structure of Bifidobacterium bifidum β-galactosidase in the PDB database shared the most similarities with the modeled structure when structural comparisons were made.
Discussion
The present study revealed that Lactiplantibacillus isolates may be a promising source of β-galactosidase enzyme. The isolate stabilized throughout a pH range of 4 to 9 and exhibited the highest β-galactosidase activity at pH 7. The different types and quantities of carbon and nitrogen sources in the culture media play a crucial role in β-galactosidase production. Gomaa (17) reported that the maximum output of β-galactosidase was produced utilizing wheat flour as the carbon source for the β-galactosidase synthesis by Lactobacillus delbrueckii and Lactobacillus reuteri. Lactobacillus delbrueckii generated the most enzymes from beef extract (24.25 U/ml), while Lactobacillus reuteri produced the most β-galactosidase (20.20 U/ml) when peptone was present, while β-galactosidase activity was reported at 40˚C and pH 7. Nguyen et al (18) demonstrated that the optimal pH range for lactose hydrolysis and the temperature for ONPG β-galactosidase activity were 6.5-8 and 55˚C, respectively. Genetic variations among distinct enzyme-producing microbial strains may account for the variations in their dietary needs (19). It was evident that certain biochemical molecules are perhaps essential for the high-level production of this enzyme. A number of biomolecules, such as amino acids, nucleic acids and enzymes are produced by a vast group of microorganisms by the metabolism of both inorganic/organic forms nitrogen (20). β-galactosidase is a tetrameric enzyme with four identical subunits and a molecular weight of 116 kDa (21). In the present study, using the SDS-PAGE technique, the β-galactosidase enzyme exhibited an intense ~116 kDa protein band, which is in agreement with the findings of other studies. Elmira et al (1) demonstrated that the 116 kDa protein band was found in Lactobacillus delbrueckii, using SDS-PAGE. Nguyen et al (18), using gel electrophoresis, revealed that the enzyme was a heterodimer comprised of two subunits of 35 and 72 kDa and the activity of β-galactosidase was 158 U/ml extracted from Lactobacillus reuteri L103 compared with Lactobacillus casei subsp. rhamnosus (22.7 U/ml). Elmira et al (1) demonstrated that the Lactobacillus delbrueckii had the highest enzymatic value (1,966 U/ml). Furthermore, a pH 7 has been shown to be ideal for mesophilic β-galactosidase production (22). In the present study, the Km value of β-galactosidase activity was found to be 27.37 mM, estimated from L. plantarum GV54 compared with other values.
Gomaa (17) also reported that the optimum β-galactosidase activity was recorded at 15 mM ONPG at 40˚C and pH 7. The enzyme activity was further enhanced in the presence of Mg2+, Mn2+and Fe2+, while it was decreased by Ag+ and Ni+ for both the Lactobacillus delbrueckii and Lactobacillus reuteri strains. In the present study, Mn2+ was shown to have the highest relative enzymatic activity, and the addition of Mg2+, Mn2+ and Fe2+ had an indirect effect on the relative enzymatic activity. The enzyme activity was nonetheless reduced by the presence of Ag+. In the present study, using the Lineweaver-Burk plot, the kinetic parameters of β-galactosidase using the ONPG substrate were determined. It was found that the Km was 27.3757 mM and the Vmax was 0.2592 U/min. Nguyen et al (18) deemonstrated that, in terms of D-glucose released per minute per mg of protein, lactose and O-nitrophenol had Km and Vmax values of 4.04±0.26 mM and 28.8±0.2 mmol, and 0.73±0.07 mM and 361 12 mmol, respectively. Di Serio et al (23) demonstrated the kinetics of β-galactosidase from Kluyveromices marxianus lactis, and a kinetic model based on the Michaelis-Menten mechanism was created, and the associated parameters were calculated.
β-galactosidase is used as a biosensor to estimate lactose present in milk and in the production of lactose-hydrolyzed milk for the treatment of lactose intolerance (8,9). Bioinformatics is an essential tool for the construction of the β-galactosidase structure, identification, localization, function and modification (12). In the present study, the 3D structure of β-galactosidase was constructed by amino acid sequences retrieved from L. plantarum exhibiting maximum resemblance with the structure of β-galactosidase from Bifidobacterium bifidum. This led to the obvious conclusion that the functional range of the L. plantarum enzyme perhaps expanded. Additionally, it has been well-documented that the enzyme in L. plantarum functions as a ligand for β-D-galactose. Ghosh et al (12) reported that the high-resolution experimental structure of the same enzyme from Penicillium sp. was similar to the modeled structure of β-galactosidase from A. niger. Thus, it can be inferred that the functioning of the A. niger enzyme may be expanded. Additionally, the enzyme in A. niger responds to β-D-galactose as a ligand (12). The modeled structure had five domains that were located in various residue locations. In the first domain of the modeled structure, Glu200 and Glu298 were found to be two catalytic residues. The structure was molecularly docked with β-D-galactose as well (12).
Bifidobacterium bifidum is widely used as a commercial probiotic in the food industry. The ability of this bacterial group to utilize short-chain β-galactosides, such as galacto-oligosaccharides (GOS) as an energy source is a key factor for the upregulation of the bacterial population levels in the human intestine (14). In fact, >8% of the genome of Bifidobacteria is related to carbohydrate metabolism; however, the molecular level understanding of the complexity of their metabolism remains elusive. In the process of hydrolysis, a water molecule plays the role of the electron acceptor necessary for break the covalent bond. Eventually, the acceptor of the reaction can also be the hydroxyl group of another sugar, such as glucose, for example, leading to the formation of GOS (22). The latter saccharides are classified as prebiotic sugars due to their ability to promote the growth of beneficial bacteria in the gut (9). In the human gastrointestinal tract, GOS are the fermentable substrates for probiotic organisms. Bifidobacteria compose almost 4.5% of the total gastrointestinal microbiota, and are known for their ability to utilize GOS, fructooligosaccharides, pectin and mucin as substrates (14). The crystal structures of the GH42 β-galactosidase BbgII from Bifidobacterium sp, a probiotic bacteria, for D-α-galactose, docking, as well as molecular dynamics simulations have elucidated that the enzyme is recognized, specifically and tightly bound to D-α-galactose (14). Using these data, the 3D structure of the amino acid sequence of β-galactosidase from L. plantarum was constructed. Considering the aforementioned characteristics, L. plantarum GV54 was considered the most efficient β-galactosidase producer for food and dairy technological applications. However, as the structure of β-galactosidase is predictive and not yet experimentally determined, there is thus a need to determine the protein sequence analysis of L. plantarum GV54.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MV executed the experimental work, prepared the manuscript and was involved in the interpretation and analysis of the data. DG conceptualized and designed the study, and was involved in the acquisition, analysis, or interpretation of data for the study. DG drafted the manuscript and revised it critically for important intellectual content. MV and DG confirm the authenticity of all the raw data. Both authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Elmira G, Fariba H, Bahareh KS, Jamileh N and Farahnaz M: Study on β-galactosidase enzyme produced by isolated lactobacilli from milk and cheese. Afri J Microbiol Res. 4:454–458. 2010. | |
|
Balamurugan R, Chandragunasekaran AS, Chellappan G, Rajaram K, Ramamoorthi G and Ramakrishna BS: Probiotic potential of lactic acid bacteria present in homemade curd in southern India. Indian J Med Res. 140:345–355. 2014.PubMed/NCBI | |
|
Jurado E, Camacho F, Luzon G and Vicaria JM: A new kinetic model proposed for enzymatic hydrolysis of lactose by a β-galactosidase from Kluyveromyces fragilis. Enzyme Microb Technol. 31:300–309. 2002. | |
|
Sriphannam W, Lumyong S, Niumsap P, Ashida H, Yamamoto K and Khanongnuch C: A selected probiotic strain of Lactobacillus fermentum CM33 isolated from breast-fed infants as a potential source of β-galactosidase for prebiotic oligosaccharide synthesis. J Microbiol. 50:119–126. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Heyman MB: The committee on nutrition. Lactose intolerance in infants, children, and adolescents. J Pediatrics. 118:1297–1286. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Oliveira C, Guimarães PM and Domingues L: Recombinant microbial systems for improved β-galactosidase production and biotechnological applications. Biotechnol Adv. 29:600–609. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Vasiljevic T and Jelen P: Production of β-galactosidase for lactose hydrolysis in milk and products using thermophilic lactic acid bacteria. J Innov Food Sci Emerg Technol. 2:75–85. 2001. | |
|
Staiano M, Bazzicalupo P, Rossi M and D'Auria S: Glucose biosensors as models for the development of advanced proteinbased biosensors. Mol Biosyst. 1:354–362. 2005.PubMed/NCBI View Article : Google Scholar | |
|
Mlichova´ Z and Rosenberg M: Current trends of β-galactosidase application in food technology. J Food Nutr Res. 2:47–54. 2006. | |
|
Selvarajan E and Mohanasrinivasan V: Kinetic studies on exploring lactose hydrolysis potential of β galactosidase extracted from Lactobacillus plantarum HF571129. J Food Sci Technol. 52:6206–6217. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Holm L and Rosenström P: Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Ghosh SK, Pandey A, Arora S and Dwivedi VD: Comparative modeling and docking studies of β-galactosidase from Aspergillus niger. Netw Model Anal Health Inform Bioinform. 2:297–302. 2013. | |
|
Dwivedi VD, Arora S and Pandey A: Computational analysis of physico-chemical properties and homology modeling of carbonic anhydrase from Cordyceps militaris. Netw Model Anal Health Inform Bioinforma. 2:209–212. 2013. | |
|
Godoy AS, Camilo CM, Kadowaki MA, Muniz HD, Santo MS, Murakami MT, Nascimento AS and Polikarpov I: Crystal structure of β1→ 6-galactosidase from Bifidobacterium bifidum S17: trimeric architecture, molecular determinants of the enzymatic activity and its inhibition by α-galactose. FEBS J. 283:4097–4112. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Vasudha M, Prashantkumar CS, Mallika B, Vishwas K and Gayathri D: Probiotic potential of β-galactosidase-producing Lactic Acid Bacteria for Lactose intolerance from fermented milk and their molecular characterization. Biomed Rep (In Press). | |
|
Vinderola CG and Reinheimer JA: Lactic acid starter and probiotic bacteria: A comparative ‘in vitro’ study of probiotic characteristics and biological barrier resistance. Food Res Int. 36:895–904. 2003. | |
|
Gomaa EZ: β-galactosidase from Lactobacillus delbrueckii and Lactobacillus reuteri: Optimization, characterization and formation of galactooligosaccharides. Indian J Biotechnol. 17:407–415. 2018. | |
|
Nguyen TH, Splechtna B, Steinböck M, Kneifel W, Lettner HP, Kulbe KD and Haltrich D: Purification and characterization of two novel β-galactosidases from Lactobacillus reuteri. J Agric Food Chem. 54:4989–4998. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Hall AB, Tolonen AC and Xavier RJ: Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 18:690–699. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Akcan N: High-level production of extracellular β-galactosidase from Bacillus licheniformis ATCC 12759 in submerged fermentation. Afr J Microbiol Res. 5:4615–4621. 2011. | |
|
Białkowska AM, Cieśliński H, Nowakowska KM, Kur J and Turkiewicz M: A new β-galactosidase with a low temperature optimum isolated from the Antarctic Arthrobacter sp. 20B: Gene cloning, purification and characterization. Arch Microbiol. 191:825–835. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Iqbal S, Nguyen TH, Nguyen HA, Nguyen TT, Maischberger T, Kittl R and Haltrich D: Characterization of a heterodimeric GH2 β-galactosidase from Lactobacillus sakei Lb790 and formation of prebiotic galacto-oligosaccharides. J Agric Food Chem. 59:3803–3811. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Di Serio M, Maturo C, De Alteriis E, Parascandola P, Tesser R and Santacesaria E: Lactose hydrolysis by immobilized β-galactosidase: The effect of the supports and the kinetics. Catalysis Today. 79:333–339. 2003. |