Caffeic acid phenethyl ester: Unveiling its potential as a potent apoptosis inducer for combating hypopharyngeal squamous cell carcinoma
- Authors:
- Published online on: December 11, 2023 https://doi.org/10.3892/or.2023.8680
- Article Number: 21
-
Copyright: © Kim et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Hypopharyngeal squamous cell carcinoma (HSCC) is a type of cancer that originates in the cells lining the bottom of the throat, known as the hypopharynx, which connects the esophagus to the larynx. Although this type of cancer represents only a small fraction of all head and neck cancers, its prognosis is markedly worse compared with other types of head and neck cancer, characterized by an overall 5-year survival rate of ~30 to 35% (1). There are three main treatment options for hypopharyngeal cancer: i) Surgery, ii) radiotherapy and iii) chemotherapy; however, the optimal therapy regimen depends mainly on the stage of the cancer. Recently, a multidisciplinary regimen involving surgery or radiotherapy with chemotherapy simultaneously has been recommended to treat advanced HSCC (2). Due to its specific localization near the larynx, chemoradiotherapy is preferred over surgical resection with adjuvant chemotherapy, to preserve laryngeal functions, including breathing, swallowing and speaking (2). In this context, regardless of the selected regimen, in HSCC, chemotherapy can be beneficial for shrinking tumor lesions, mitigating locoregional recurrence and improving disease-free survival (3), thereby highlighting the need for novel chemotherapeutic drugs with high efficacy.
Caffeic acid phenethyl ester (CAPE) is the primary biologically active phenolic compound found in honeybee propolis and has been documented to have anti-inflammatory, antioxidant, antiviral and anti-microbial features (4). CAPE has also been identified as a prospective anticancer drug candidate due to its selective cytotoxicity toward cancer cells (5). Its therapeutic versatility has been demonstrated based on its efficacy as a chemotherapeutic adjuvant, as it potentiates the effectiveness of therapy while mitigating the chemotherapy-induced side effects in various cancers (6). Moreover, CAPE treatment also inhibits the proliferation, survival and invasion of oral cancer cells by inhibiting various signaling pathways, such as EGFR, Akt and NF-κB signaling (7–9). However, further exploration of the antitumor effects of CAPE specifically on hypopharyngeal cancer remains necessary.
Inhibitors of apoptosis proteins (IAPs) comprise a family of proteins that controls cell death. Survivin and X-linked IAP (XIAP) are the essential members of the IAP family; they exert inhibitory effects on caspase-activity and are recognized as promising therapeutic targets of cancer therapy due to their overexpression in a variety of cancers (10,11). Survivin, which is well known for its ability to inhibit apoptosis and regulate the cell cycle, exhibits high expression in most cancers and is associated with tumor aggressiveness and unfavorable clinical outcomes (12). Furthermore, the expression of survivin has been linked to a poor prognosis and short overall survival, rendering it a possible diagnostic marker for head and neck squamous cell carcinoma (13,14). XIAP is the most powerful member of the IAP family, as it directly interacts with, and counteracts caspases 3, 7 and 9 (15). It has been documented that XIAP expression appears to represent a negative predictor of prognosis in head and neck cancer (16) and in laryngeal carcinoma (17). Moreover, survivin and XIAP can be combined to form a complex known as IAP-IAP, in which they work together to counteract the activity of caspase-9 (18). Thus, it may be a promising approach to develop novel medications that concurrently target survivin and XIAP during cancer therapy.
In the present study, the pharmacological activity of CAPE regarding the induction of intrinsic mitochondrial apoptosis, which was ascribed to the concurrent inhibition of the survivin and XIAP proteins was assessed, suggesting that CAPE is a potent drug candidate in the therapy of HSCC.
Materials and methods
Cell culture and reagents
The FaDu and SNU-1041 human HSCC cell lines were purchased from the Korean Cell Line Bank (Seoul, Republic of Korea) and cultured in either Minimum Essential Medium (FaDu) or RPMI-1640 medium (SNU-1041) from Welgene, Inc. containing 10% FBS (Welgene, Inc.) and 1% penicillin/streptomycin (P/S). The IHOK cell line was kindly provided by Yonsei University (Seoul, Republic of Korea) and cultured in KBM™ Gold Keratinocyte Growth Basal Medium (Lonza Group, Ltd.) supplemented with KGMTM-2 Keratinocyte Growth Medium-2 SingleQuots™ Supplements and Growth Factors (Lonza Group, Ltd.). The cells were incubated at 37°C in a humidified environment with 5% CO2. All experiments were conducted when the cells had grown to ~50% confluence. All chemicals were dissolved in DMSO and stored at −20°C. CAPE was purchased from Santa Cruz Biotechnology, Inc. Each cell line was divided into two groups for further analysis of cellular responses to CAPE treatment: i) The vehicle control group and ii) the CAPE (70 µM for FaDu and 100 µM for SNU-1041 and IHOK) treatment group. Vehicle- and CAPE-treated cells were incubated at 37°C for 24 h. For pre-treatment of inhibitors for 1 h (Z-VAD-FMK) or 2 h (CHX and MG132) at 37°C, each cell line was divided into following two groups: i) Vehicle control group and ii) Z-VAD-FMK (10 µM), CHX (100 ng/ml for FaDu and 50 ng/ml for SNU-1041), and MG132 (800 nM) treatment group.
Trypan blue assay
FaDu, SNU-1041 and IHOK cells at 50% confluency were treated with vehicle or CAPE. Trypan blue solution in a concentration of 0.4% (Corning, Inc.) was used to stain the cells at a 1:1 ratio at room temperature (RT). Immediately, after staining, cell viability was evaluated automatically using a CytoSMART cell counter (Corning, Inc.).
Soft agar assay
Basal Medium Eagle (BME; Sigma-Aldrich,) was dissolved in distilled water (DW) supplemented with sodium bicarbonate, resulting in a 2X BME solution, then filtered using a 0.2-µm filter (Sartorius AG). A mixture containing 1.25% agar, 2X BME, FBS, PBS, l-glutamine and gentamicin was then prepared. A total of 3 ml of the 1.25% agar mixture containing the indicated amounts of CAPE (70 µM for FaDu and 100 µM for SNU-1041) was added to each well of the 6-well plates. The agar mixture was then allowed to solidify for 2 h at RT. Subsequently, a 10% BME solution was prepared by diluting 2X BME with DW and supplementing with l-glutamine, gentamicin and FBS. FaDu and SNU-1041 cells were suspended in the 10% BME solution and mixed with 1.25% agar. After mixing, 1 ml of the mixture containing 0.8×104 cells was added directly onto the 6-well solid-bottom agar plates. The plates were incubated at RT for 2 h for agar solidification and then placed in a humidified incubator at 37°C with 5% CO2 for a period of ~4 weeks. The 6-well plates were supplemented once a week with 150 µl of complete medium containing the indicated concentrations of CAPE, to prevent agar desiccation. Images of cell colonies were captured using a CKX53 microscope (Olympus Corporation) and colony counts were analyzed automatically using the ImageJ software, version 1.53t (National Institutes of Health).
Sphere formation assay
Cells were suspended in a serum-free medium supplemented with 0.01X N-2 and B-27 supplements, 25 ng/ml of human basic fibroblast growth factor (bFGF; Invitrogen; Thermo Fisher Scientific, Inc.) and human epidermal growth factor (EGF; Thermo Fisher Scientific, Inc.) and 1% P/S. The cells (1×104 cells/well) were cultured in ultralow attachment 6-well plates (Corning, Inc.) for 5 days (FaDu) and 11 days (SNU-1041). Spheres were randomly photographed using an inverted light microscope (Nikon Corporation) and counted automatically using ImageJ software, version 1.53t.
Evaluation of nuclear morphological changes
FaDu and SNU-1041 cells, which were treated with either a vehicle or CAPE when they reached ~50% confluence were fixed with 70% ethanol at −20°C overnight and stained with a DAPI solution (2 µg/ml; MilliporeSigma) on a glass slide for 10 min at RT. Images of the nuclear morphological changes occurring in apoptotic cells were captured by fluorescence microscopy (Leica DMi8; Leica Microsystems GmbH).
Measurement of cell-cycle distribution
After treating the cells with vehicle or CAPE when they reached ~50% confluence, FaDu and SNU-1041 cells were fixed with 70% ethanol overnight at −20°C and stained with a propidium iodide (PI) solution (20 µg/ml; MilliporeSigma) containing RNase A (20 µg/ml; Thermo Fisher Scientific Inc.) at 37°C for 15 min. Cell-cycle distribution was then assessed by an LSRFortessa X-20 flow cytometer (BD Biosciences) and analyzed using the FlowJo software, version 10.8.1 (FlowJo LLC).
Determination of apoptotic cell populations
FaDu and SNU-1041 cells, which were treated with either a vehicle or CAPE when they reached approximately 50% confluence were stained with FITC Annexin V Apoptosis Detection Kit (cat. no. 556547; BD Pharmingen; BD Biosciences), according to manufacturer's instructions. In brief, the cells were stained with Annexin V for 15 min at RT and then stained with PI on ice just before evaluation using a flow cytometer. The apoptotic population was evaluated using an LSRFortessa X-20 flow cytometer (BD Biosciences) and analyzed using the FlowJo software, version 10.8.1.
Western blot analysis
Total protein lysates were extracted using 1X RIPA buffer (MilliporeSigma) including cOmplete™, Mini protease inhibitor cocktails (Roche Diagnostics) and Pierce™ phosphatase inhibitor tablets (Thermo Scientific; Thermo Fisher Scientific, Inc.). A quantitative analysis of protein concentrations was carried out using a DC Protein Assay Kit (Bio-Rad Laboratories, Inc.). Equal amounts of protein were loaded in each lane of the gel, but this amount was adjusted according to the protein examined. Protein lysates were separated using SDS-PAGE. The separated proteins were then electrotransferred onto a PVDF membrane. The membrane was blocked with 5% skim milk in TBST including 0.05% Tween 20 (Duchefa Biochemie) for 1 h 30 min at RT, to prevent non-specific binding of antibodies to the membrane. Subsequently, the membrane was thoroughly rinsed with TBST, incubated with the designated primary antibodies overnight at 4°C, then exposed to HRP-conjugated secondary antibodies for 4 h at 4°C. The protein bands of interest were visualized using the WestGlow™ PICO PLUS chemiluminescent Substrate (BIOMAX) on an X-ray film or using the Image Quant LAS 500 system (Cytiva). β-actin was used as a loading control. The intensity of protein bands was quantified using ImageJ software, version 1.53t. Information concerning the antibodies used as well as the amount of loaded protein and the percentages of the gels for the experiments conducted are presented in Table I.
Assessment of changes on the mitochondrial membrane potential (ΔΨm)
The effects of CAPE on the ΔΨm were determined using a MitoScreen JC-1 kit (cat. no. 551302; BD Pharmingen; BD Biosciences), according to the manufacturer's protocols. Briefly, FaDu and SNU-1041 cells were washed twice with PBS and incubated with a 1X JC-1 working solution for 30 min at 37°C. The stained cells were washed and resuspended in 1X assay buffer, then analyzed using an LSRFortessa X-20 flow cytometer (BD Biosciences) and interpreted using the FlowJo software, version 10.8.1.
Cytosolic and mitochondrial fractionation
Vehicle- or CAPE-treated FaDu and SNU-1041 cells were separated into cytosolic and mitochondrial extracts using the Mitochondria/Cytosol Fractionation Kit (cat. no. ab65320; Abcam), according to the manufacturer's protocols. In brief, the cells were resuspended in 1X cytosol extraction buffer mix including protease inhibitors and DTT, vortexed for 2 min and placed on ice for 10 min, to allow complete disruption of cell membranes. Following centrifugation for 15 min at −10,000 × g at 4°C, the supernatants (including the cytosolic fraction of the cells) were carefully transferred to new microtubes. The remaining cell pellets were washed with PBS, resuspended in 1X mitochondrial extraction buffer mix containing protease inhibitors and DTT, vortexed for 10 sec, and centrifuged using the aforementioned conditions. The resulting supernatants (including the mitochondrial fraction of the cells) were collected in new microtubes.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was utilized to extract total RNA from FaDu and SNU-1041 cells and 1 µg of the extracted RNA was reverse transcribed into cDNA using the AMPIGENE cDNA Synthesis Kit (Enzo Life Sciences, Inc.): 42°C for 30 min and 85°C for 10 min. The resulting cDNA was used as the input for PCR using the AMPIGENE qPCR Green Mix Hi-Rox reagent (Enzo Life Sciences, Inc.). Real-time PCR was carried out on an Applied Biosystems StepOne Plus Real-Time PCR System (Applied Biosystems) using the following conditions for all genes: 95°C for 2 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec. The amount of the GAPDH gene was used to normalize the relative amount of each gene, which was measured using the 2−ΔΔCq method (19). The qPCR primers used in this experiment were as follows: Survivin forward, 5′-ACTTGGCCCAGTGTTTCTT-3′ and reverse, 5′-GACAGAAAGGAAAGCGCAAC-3′; XIAP forward, 5′-TGGTATCCAGGGTGCAAATATC-3′ and reverse, 5′-GTTCTTACCAGACACTCCTCAAG-3′; and GAPDH forward, 5′-GTGGTCTCCTCTGACTTCAAC-3′ and reverse, 5′-CCTGTTGCTGTAGCCAAATTC-3′. The primers were designed based on the NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) reference sequences: Survivin-CR541740.1, XIAP-BC032729.1 and GAPDH-BC083511.1. The detailed primer binding sites of respective proteins are included in the Fig. S1, Fig. S2, Fig. S3.
Statistical analysis
All graphs were generated using GraphPad Prism, version 8.0 (GraphPad Software; Dotmatics) and all statistical analyses were performed using SPSS, version 26.0 (IBM, Corp). The results of three independent biological experiments are presented as the average (mean) value together with the measure of variation [standard deviation (SD)] for all data. Statistical significance was determined by conducting either an unpaired two-tailed Student's t-test or a one-way ANOVA with Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
CAPE inhibits the anchorage-dependent and independent growth of HSCC cell lines
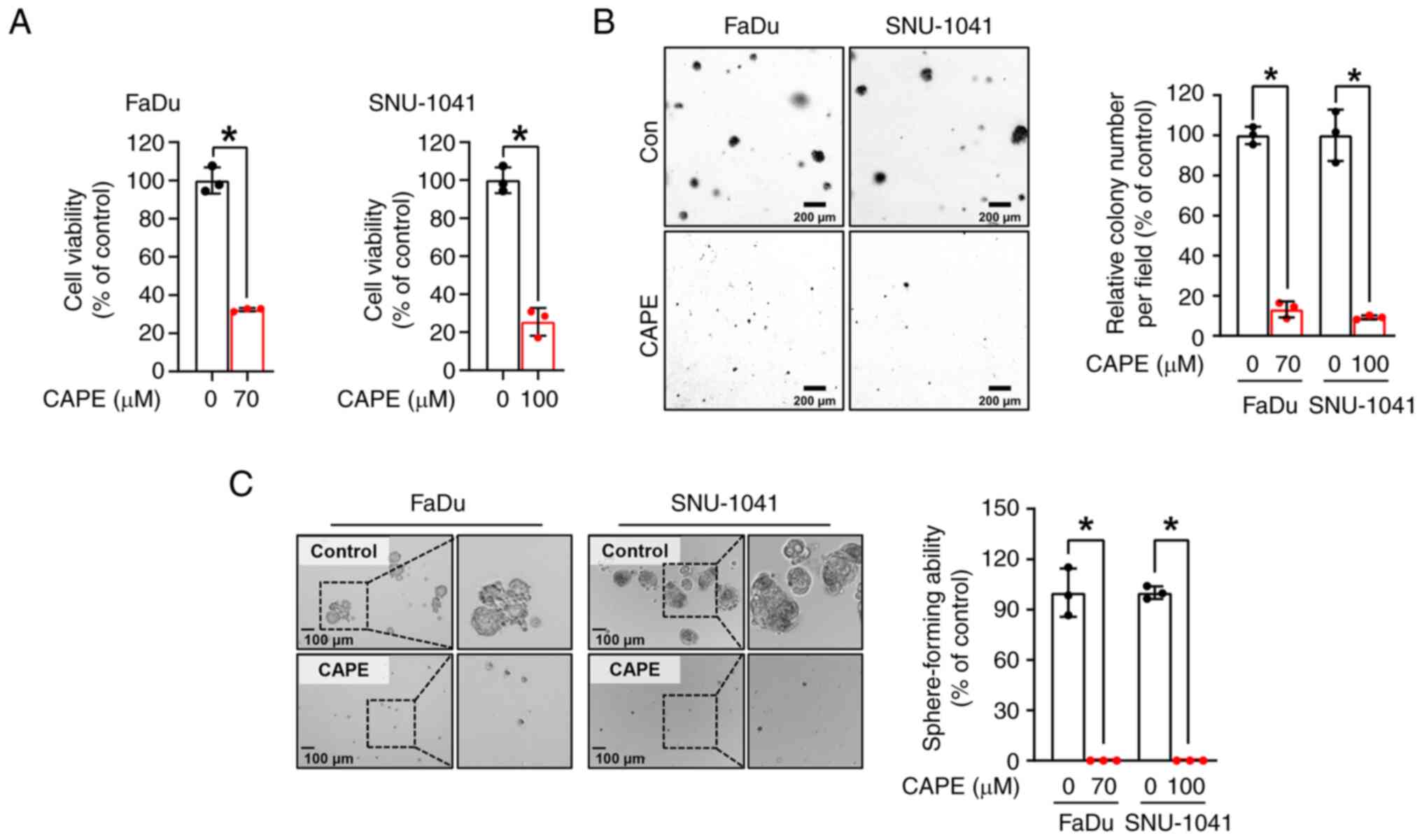
To examine the impacts of CAPE on HSCC cell growth, a trypan blue assay was performed on two HSCC cell lines, FaDu and SNU-1041, which had been treated with CAPE for 24 h. As revealed in Fig. 1A, the growth of the two cell lines was significantly reduced after treatment with CAPE, but not with the vehicle control. Notably, it was observed that CAPE worked less effectively in the IHOK cell line, immortalized human oral keratinocytes, compared with HSCC cell lines, suggesting that CAPE could selectively attenuate the growth of cancer cells rather than affect normal cells (Fig. S4). A soft agar assay was then conducted to further evaluate the influence of CAPE on the anchorage-independent growth of the HSCC cell lines. As displayed in Fig. 1B, the clonogenic activity of the two cell lines was attenuated by CAPE treatment, as indicated by the reduced size and number of the cell colonies. Furthermore, to examine the tumor suppressive capability of CAPE in a 3D culture system, a sphere-formation assay was carried out. As demonstrated in Fig. 1C, the sphere-forming ability of both cell lines was strongly inhibited in CAPE-treated cells compared with control cells. These findings suggested that CAPE thwarts cell growth and carcinogenesis in these HSCC cell lines.
CAPE impedes tumorigenicity by triggering caspase-dependent apoptosis in HSCC cell lines
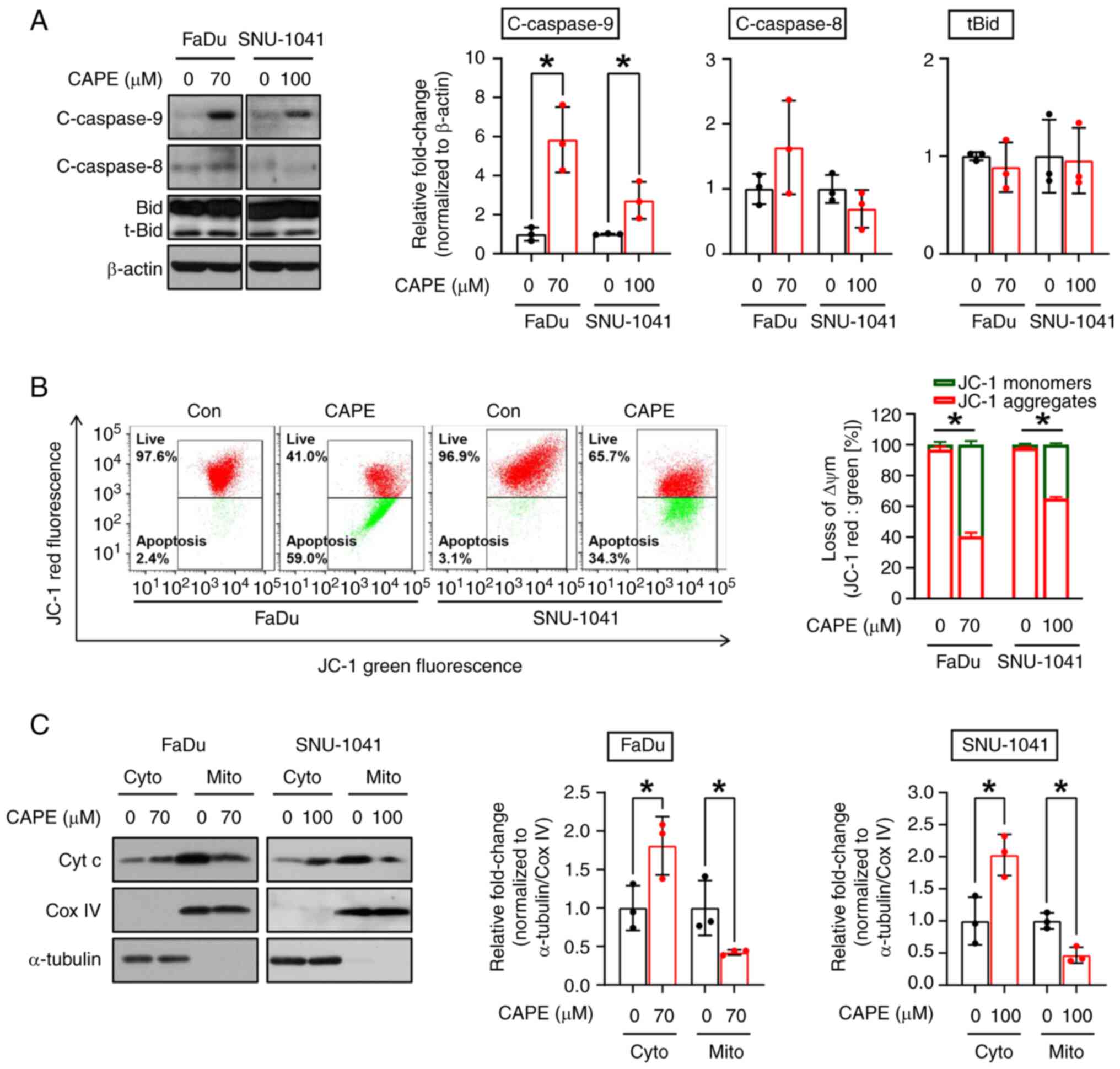
To confirm whether the observed cytotoxic effect of CAPE on HSCC cell lines was caused by the activation of apoptotic signaling, three apoptosis detection assays were employed. First, the morphological changes in the nuclei of CAPE-treated cells compared with vehicle-treated cells were evaluated; fragmented or condensed nuclei were observed, which are regarded as typical morphological signs of apoptosis (Fig. 2A). Subsequently, whether cell populations within the sub-G1 phases expanded after CAPE treatment was investigated. As illustrated in Fig. 2B, a significant increase in the sub-G1 population in both cell lines was observed after the application of CAPE. Finally, apoptotic cell populations were measured using annexin/PI double staining, to investigate further the effect of CAPE on the induction of apoptosis. It was also revealed that CAPE treatment enhanced the populations within the early and late apoptotic compartments (Fig. 2C). Based on these results, whether the apoptosis stimulated in the cells exposed to CAPE was dependent on the activation of caspase signaling, was examined. As shown in Fig. 3A, the CAPE treatment notably stimulated the formation of cleaved forms of caspase-3 (c-caspase-3) and induced the expression of cleaved poly(ADP-ribose) polymerase (c-PARP). To obtain evidence to corroborate these results, the cells were treated with Z-VAD-FMK, which is a caspase-inhibitor, for 1 h before exposing them to CAPE treatment for 24 h, to investigate whether CAPE can induce caspase-dependent apoptosis in the two cell lines. As expected, the induction of the expression of c-caspase-3 and c-PARP and annexin V-positive compartments by the CAPE treatment was reversed by pre-treatment of the cells with Z-VAD-FMK (Fig. 3B and C). Taken together, these results indicated that CAPE considerably attenuates the growth of HSCC cells, possibly by triggering caspase-dependent apoptotic processes.
CAPE exerts cell-killing effects through the activation of mitochondrial apoptotic signaling in HSCC cell lines
Apoptosis can be triggered via two primary pathways, the extrinsic or intrinsic pathway, with formation of cleaved caspase-8 and caspase-9 being considered a typical marker of each pathway, respectively. Therefore, the pathway which was responsible for the apoptotic cell-killing effect in cells that were subjected to CAPE treatment was investigated. After exposure of both cell lines to CAPE for 24 h, an increase in the expression of cleaved caspase-9 (c-caspase-9), but not of cleaved caspase-8 (c-caspase-8), Bid, and t-Bid were observed, indicating that CAPE may promote intrinsic apoptosis signaling (Fig. 4A). In the intrinsic pathway, mitochondria lose their ΔΨm and exhibit increased membrane permeabilization, which causes the release of proteins from the mitochondrial intermembrane spaces, such as cytochrome c (20). To demonstrate further the impact of CAPE on the activation of the intrinsic apoptotic pathway, changes in ΔΨm were measured by employing the JC-1 fluorescent dye. As expected, the number of JC-1 aggregates (red fluorescence) was decreased in cells treated with CAPE, indicating that the treatment caused a reduction in the ΔΨm (Fig. 4B). Next, it was investigated whether the changes in the ΔΨm caused by CAPE led to the outer mitochondrial membrane permeabilization, causing the release of cytochrome c. Cytochrome c oxidase subunit 4I1 (Cox IV), a cytochrome c oxidase enzyme located within the inner membrane of the mitochondria (21), and α-tubulin, a cytoplasmic marker protein (22), were used as mitochondrial and cytosol loading controls, respectively. As demonstrated in Fig. 4C, it was found that cytochrome c was transferred from the mitochondria to the cytosol, as indicated by the decrease in cytochrome c detected in the mitochondria and the increase in cytochrome c observed in the cytosol in response to CAPE. Accordingly, the apoptosis observed in the two CAPE-treated cell lines was attributed to an intrinsic mitochondrial signaling process.
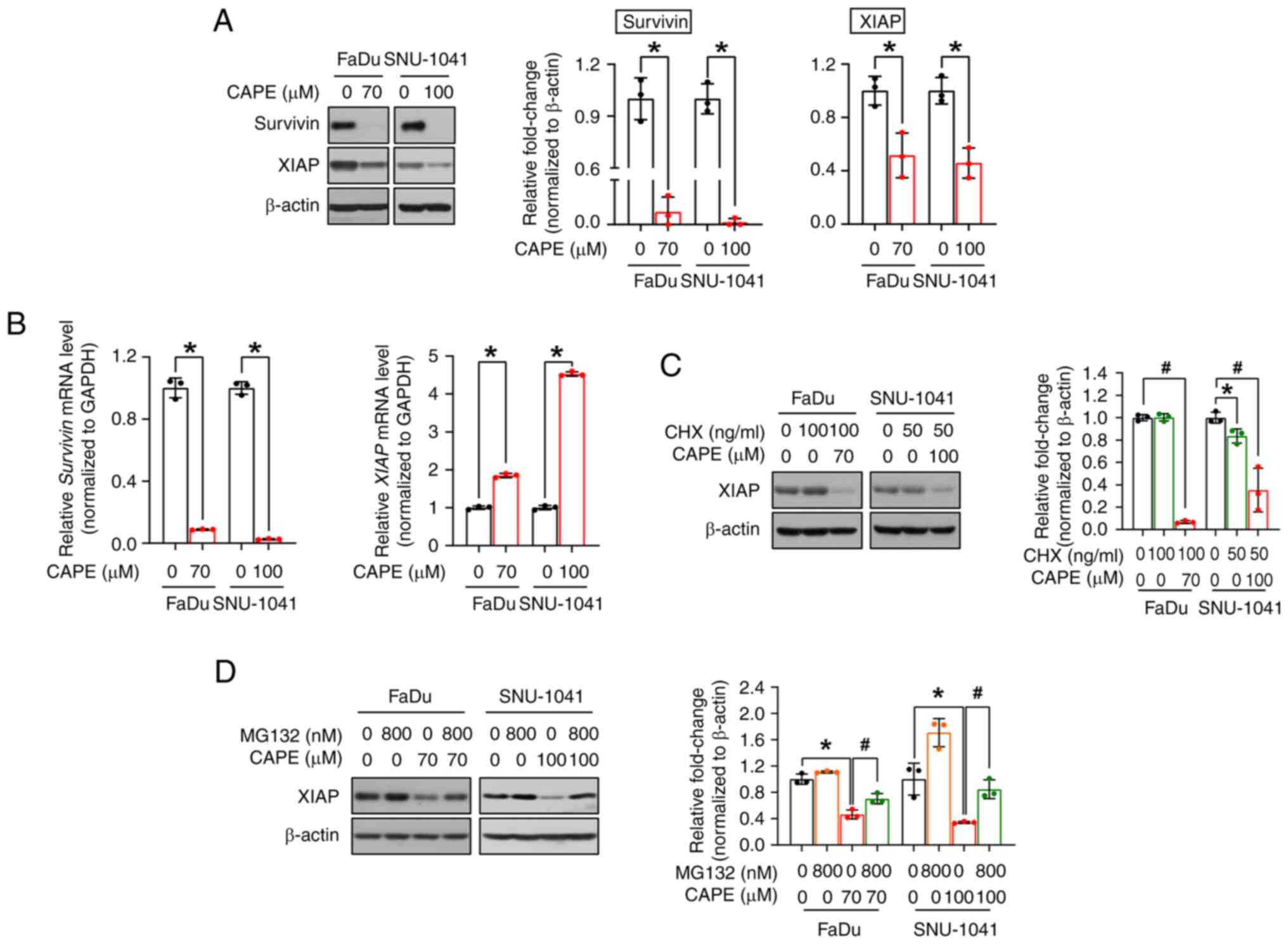
CAPE induces an apoptotic signaling cascade by blocking the anti-apoptotic proteins survivin and XIAP
As aforementioned, survivin and XIAP are the major members of the IAP family, and both hinder apoptosis, leading to the increased survival of cancer cells (23). Therefore, the expression of survivin and XIAP were investigated to determine the exact mechanism underlying CAPE-mediated apoptosis. As expected, CAPE administration resulted in the downregulation of both proteins (Fig. 5A). Next, to investigate whether the changes observed for the two proteins were caused by transcriptional regulation, the corresponding mRNA expression levels were measured. As revealed in Fig. 5B, CAPE downregulated the survivin mRNA, but not the XIAP mRNA, suggesting that survivin alone was regulated at the transcriptional level. Therefore, it was hypothesized that XIAP is regulated at the post-translational level, and it was investigated whether CAPE compromises the stability of this protein. In advance of implementing cycloheximide (CHX) and MG132 experiments, a time-course experiment was conducted to verify the exact time point of XIAP degradation by CAPE and to minimize its potential effect on cell viability. In Fig. S5, XIAP was downregulated after 6 h of CAPE treatment in both cell lines. The time was determined when a 1-h pretreatment of inhibitors was followed by subsequent CAPE treatment for 6 h. As depicted in Fig. 5C, the level of the XIAP protein was decreased in the two cell lines after co-treatment with CHX (a protein synthesis inhibitor) and CAPE, which may indicate that XIAP was controlled by post-translational regulation, particularly via protein degradation. Thus, the cells were subsequently treated with MG132 (a proteasome inhibitor) for 2 h prior to CAPE treatment, to explore whether XIAP protein degradation was accelerated by CAPE in a proteasome-dependent manner. In Fig. 5D, MG132 treatment restored the downregulated levels of expression of the XIAP protein in the CAPE-treated groups in both cell lines. Consequently, these data indicated that CAPE exerts its apoptosis-inducing effects on HSCC cells by downregulating two anti-apoptotic proteins, survivin and XIAP, via either transcriptional or post-translational regulation.
Discussion
In the present study, the potential of the bioactive component CAPE to treat HSCC via the induction of mitochondrial apoptosis was demonstrated and was achieved by blocking two potent anti-apoptotic proteins, survivin and XIAP.
In the present study, cells were initially treated with both 100 and 120 µM of CAPE for 24 h and it was observed that 120 µM of CAPE significantly decreased the expression of β-actin indicating excessive cytotoxicity (Fig. S6A). Consequently, the concentrations were adjusted to a range of 60–100 µM, ensuring that the expression of β-actin remained constant while c-PARP increased suitably (Fig. S6B). While a higher CAPE dose could induce more prominent apoptotic effects, it also carried a risk of causing excessive cytotoxicity in both normal and cancer cells. Based on the aforementioned data, 70 µM CAPE for FaDu and 100 µM CAPE for SNU-1041 were employed, to demonstrate its anticancer therapeutic potential, while avoiding any significant effect on the expression of β-actin. Several previous studies have employed varying concentrations of the same drug for different cell lines in vitro (24–27). In the case of CAPE, CAPE was administered at concentrations of 10–50 µM to multiple myeloma cell lines to investigate its antimyeloma potential (28). A different study exposed human colorectal cell lines to 75 µM CAPE, revealing CAPE-induced apoptotic cell death through the inhibition of survivin (29). Furthermore, ovarian cancer OV7 cells were treated with 5–100 µM of CAPE to explore its therapeutic benefits in serous ovarian cancer (30). In a separate study, endometrioid ovarian carcinoma cells were exposed to CAPE at concentrations ranging from 1–10 µM, demonstrating its anti-tumorigenic activity (31). It was also revealed that CAPE at concentrations ranging from 1–50 µM could induce apoptosis and oxidative stress in human multiple myeloma cells (32). These findings indicated that in vitro treatment concentration of some substances may be dependent on cell context even though they exhibited similar functions such as apoptosis induction.
Targeting apoptosis has been deemed a successful approach in cancer therapy as apoptosis evasion is the hallmark of various cancers, regardless of their etiology and type (33). Apoptosis can be triggered by two primary pathways such as the binding of death ligands to their cognate death receptors, which is termed the extrinsic pathway and cytotoxicity from mitochondrial signaling, which is termed the intrinsic pathway (34). The extrinsic pathway relies on Fas, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or TNFα to transmit extracellular signals into cells, which ultimately leads to the activation of caspase-8. By contrast, in the intrinsic pathway, intercellular stimuli such as cellular stress, DNA damage and growth factor withdrawal provoke mitochondrial malfunctioning via the induction of mitochondrial outer membrane permeabilization, consequently causing the release of mitochondrial proteins, such as cytochrome c, SMA and OMI into the cytosol. Caspase-9 and apoptotic peptidase activating factor 1 (APAF1) are recruited by cytochrome c to produce cleaved caspase-9, which is the activated form of caspase-9 (35). These two apoptotic pathways eventually converge toward the formation of cleaved caspase-3 and, finally, cleaved PARP, followed by cell demise (20). Several natural products exert tumor-inhibitory activities by stimulating the intrinsic and extrinsic apoptotic signaling pathways exclusively, respectively, or by triggering the two pathways simultaneously (36–38). In the latter cellular context, the intrinsic and extrinsic apoptotic pathways can be integrated through the truncation of the BH3 interacting domain death agonist (Bid) protein, which is a process that is facilitated by c-caspase-8 (39). In the present study, it was ascertained whether CAPE exclusively triggers one type of apoptosis or whether it simultaneously induces both types of apoptosis. It was revealed that the expression of c-caspase-9 alone was increased, whereas the expression of c-caspase-8 and truncated Bid remained unchanged (Fig. 4A). Based on these results, the authors suggest that CAPE exerts its HSCC cell-destroying effects mainly via mitochondria-mediated intrinsic apoptosis.
Since apoptosis is an ongoing and meticulously controlled mechanism that sustains the cellular equilibrium within the body of a healthy animal (40), the expression of c-PARP can be detected at low levels. In Fig. 3A and B, the relative expression levels of c-PARP in vehicle- and CAPE-treated cells were measured, and basal protein expression levels were visualized due to an extended exposure time. However, it is evident that c-PARP expression levels were further induced by CAPE treatment compared to basal levels.
Survivin and XIAP are members of the IAP family that have the potential to regulate cell death, and they have been identified as reasonable targets for numerous anticancer therapeutics in various types of cancer (41–44). Although targeting these proteins individually has been shown to be an effective intervention per se, a simultaneous therapeutic intervention against the two proteins appears to be a more feasible pharmacological strategy (45–48). As aforementioned, survivin and XIAP may form a survivin-XIAP complex with a vigorous apoptosis inhibitory activity (18). Thus, targeting both survivin and XIAP, as CAPE did in the experiments of the present study, may provide improved treatment options for multiple types of cancers. In the present study, a CAPE-mediated decrease in the levels of the survivin protein was observed and appeared to be controlled by transcriptional regulation, as evidenced by the reduction of the levels of the survivin mRNA. By contrast, the mRNA level of XIAP was unexpectedly increased, whereas its protein level was significantly decreased, by CAPE treatment. Under particular cellular conditions, the expression of a given protein does not coincide perfectly with that of its cognate mRNA, possibly due to the compensatory responses of gene expression, in which the downregulation of protein levels leads to an upregulation of mRNA levels, or post-translational modification. In a recent study, it was observed that the small molecule, bufalin, induced a reduction in the expression of the E2F2 protein, whereas the expression of the E2F2 mRNA was increased, suggesting that the downregulation of the E2F2 protein was a result of post-translational modification (49). In the present study, it was also found that CAPE regulated XIAP via a post-translational modification. Survivin and XIAP complexes enhance the stability of XIAP by preventing the ubiquitination-mediated degradation of XIAP, thus resulting in the inhibition of caspase-activity followed by abnormal tumor growth (50). The binding of survivin to XIAP can also serve as a salvage mechanism by blocking the formation of the XIAP-XAF1 complex, which targets survivin for degradation through the ubiquitin-proteasome system (51). Because XIAP protein stability is partly associated with its binding to survivin, the levels of transcriptionally downregulated survivin cannot prevent the degradation of XIAP by the proteasome.
Since survivin and XIAP are regulated by different mechanisms, there may be separate upstream regulators controlling their expression, respectively. Regarding survivin, the expression decreased at the mRNA level, suggesting that transcription factors are likely associated with its downregulation. Previous studies have reported that the p53 protein could recruit itself alone or in conjunction with other proteins to the survivin promoter to suppress it (52,53). Thus, CAPE treatment possibly mediated the recruitment of p53 to the survivin promoter, ultimately reducing its expression. On the other hand, XIAP regulation appears to be executed through post-translational modifications, particularly proteasome-mediated degradation. Notably, XIAP can be phosphorylated by Akt and protein kinase C (PKC), which prevents its ubiquitination and subsequent proteasomal degradation (54,55). XIAP is also well-known for its E3 ubiquitin ligase activity, allowing it to induce autoubiquitination. Consequently, it is possible that the attenuated activation of Akt and PKCs, which cannot shield XIAP from ubiquitination, or the promotion of XIAP autoubiquitination by CAPE, could compromise its stability. Therefore, further investigation of the precise mechanisms underlying the regulation of survivin and XIAP during CAPE treatment in HSCC remains necessary.
In previous studies, in vivo experiments using CAPE have been conducted to evaluate its anticancer activity, and the results revealed that CAPE has anticancer effects in hepatocellular carcinoma and colon cancer (56,57). These findings led to the investigation of whether CAPE may also inhibit tumor growth in human HSCC in vivo. However, in the present study, the 3D sphere formation assay was implemented to demonstrate the anticancer properties of CAPE in cancer stem cell (CSC) characteristics of HSCC and to explore its potential as a therapeutic agent, as an alternative to in vivo experiments. Considering the well-recognized ability of the 3D sphere formation culture to more effectively recapitulate in vivo tumor biology compared with the 2D monolayer culture system (58), the data obtained from the 3D culture system can provide some insights that complement mouse xenograft experiments. Nevertheless, further investigations are still warranted to clarify the tumor-inhibitory effects of CAPE on HSCC in an in vivo setting.
In the clinical field, the preferred initial treatment approach, as recommended by both American and European guidelines, typically involves preserving the organ through a combination of chemotherapy and radiation therapy (59). Given this context, chemotherapy signifies much more than merely a choice for treating HSCC. The authors of the present study maintain that CAPE alone may not eliminate tumors; however, it could serve as a promising alternative to current therapeutic drugs. This, in turn, could minimize tumor regions and suppress recurrence, serving as both neoadjuvant and adjuvant chemotherapy.
Previous studies have explored the anticancer effects of CAPE on oral cancer, primarily utilizing oral squamous cell carcinoma cell lines (9,60,61). In the present study, head and neck cancer cell lines were employed, specifically FaDu and SNU-1041, which are hypopharyngeal cancer cell lines technically categorized under nasopharyngeal cancer. Although oral cancer and hypopharyngeal cancer fall within the broader classification of head and neck cancer, they possess distinct staging assessments and treatment standards due to their unique characteristics and locations within the head and neck region (62). To the best of the authors' knowledge, the present study marked the first demonstration of the cancer-inhibitory efficacy of CAPE through the induction of cell death in HSCC lines, adding novelty to the study. However, the authors used only two HSCC cell lines in this study, and further verification of the efficacy of CAPE as a cancer therapeutic for HSCC is required.
In conclusion, the present study provided initial evidence that CAPE induces the mitochondria-mediated intrinsic apoptosis pathway in HSCC by simultaneously regulating two major members of the IAP protein family. These findings suggested that CAPE offers the basis for a promising alternative strategy to cure HSCC in clinical practice.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (grant nos. 2019R1A2C1085896 and 2020R1C1C1005480).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Authors' contributions
SDC conceived and designed the study. HJK and MHA performed all the experiments, designed the primers for RT-qPCR and drafted the manuscript. JAS, SJC and HJY analyzed and interpreted the data. HJK, MHA and SDC confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Newman JR, Connolly TM, Illing EA, Kilgore ML, Locher JL and Carroll WR: Survival trends in hypopharyngeal cancer: A population-based review. Laryngoscope. 125:624–629. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Eckel HE and Bradley PJ: Treatment options for hypopharyngeal cancer. Adv Otorhinolaryngol. 83:47–53. 2019.PubMed/NCBI | |
|
Mura F, Bertino G, Occhini A and Benazzo M: Surgical treatment of hypopharyngeal cancer: A review of the literature and proposal for a decisional flow-chart. Acta Otorhinolaryngol Ital. 33:299–306. 2013.PubMed/NCBI | |
|
Pandey P, Khan F, Upadhyay TK and Giri PP: Therapeutic efficacy of caffeic acid phenethyl ester in cancer therapy: An updated review. Chem Biol Drug Des. 102:201–216. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Murtaza G, Karim S, Akram MR, Khan SA, Azhar S, Mumtaz A and Bin Asad MH: Caffeic acid phenethyl ester and therapeutic potentials. Biomed Res Int. 2014:1453422014. View Article : Google Scholar : PubMed/NCBI | |
|
Patel S: Emerging adjuvant therapy for cancer: Propolis and its constituents. J Diet Suppl. 13:245–268. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Kuo YY, Su LC, Chung CJ, Lin CY, Huo C, Tseng JC, Huang SH, Lai CJ, Chen BC, Wang BJ, et al: Caffeic Acid phenethyl ester is a potential therapeutic agent for oral cancer. Int J Mol Sci. 16:10748–1066. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Yu HJ, Shin JA and Cho SD: Inhibition of focal adhesion kinase/paxillin axis by caffeic acid phenethyl ester restrains aggressive behaviors of head and neck squamous cell carcinoma in vitro. Arch Oral Biol. 146:1056112023. View Article : Google Scholar : PubMed/NCBI | |
|
Chung LC, Chiang KC, Feng TH, Chang KS, Chuang ST, Chen YJ, Tsui KH, Lee JC and Juang HH: Caffeic acid phenethyl ester upregulates N-myc downstream regulated gene 1 via ERK pathway to inhibit human oral cancer cell growth in vitro and in vivo. Mol Nutr Food Res. 612017.doi: 10.1002/mnfr.201600842. | |
|
Frassanito MA, Saltarella I, Vinella A, Muzio LL, Pannone G, Fumarulo R, Vacca A and Mariggiò MA: Survivin overexpression in head and neck squamous cell carcinomas as a new therapeutic target (Review). Oncol Rep. 41:2615–2624. 2019.PubMed/NCBI | |
|
Tu H and Costa M: XIAP's profile in human cancer. Biomolecules. 10:14932020. View Article : Google Scholar : PubMed/NCBI | |
|
Jaiswal PK, Goel A and Mittal RD: Survivin: A molecular biomarker in cancer. Indian J Med Res. 141:389–397. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou LQ, Hu Y and Xiao HJ: The prognostic significance of survivin expression in patients with HNSCC: A systematic review and meta-analysis. BMC Cancer. 21:4242021. View Article : Google Scholar : PubMed/NCBI | |
|
Khan SA, Burke M, Zhu F, Yang DH, Dubyk C, Mehra R, Lango MJ, Ridge JA, Sher DJ and Burtness B: Survivin expression and impact on head and neck cancer outcomes. Oral Oncol. 112:1050492021. View Article : Google Scholar : PubMed/NCBI | |
|
Obexer P and Ausserlechner MJ: X-linked inhibitor of apoptosis protein-a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front Oncol. 4:1972014. View Article : Google Scholar : PubMed/NCBI | |
|
Nagi C, Xiao GQ, Li G, Genden E and Burstein DE: Immunohistochemical detection of X-linked inhibitor of apoptosis in head and neck squamous cell carcinoma. Ann Diagn Pathol. 11:402–406. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Ma X, Lu X, Cui L and Dong W: Expression of inhibitor of apoptosis protein XIAP in laryngeal carcinoma and its clinicopathological significance. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 21:973–975. 2007.(In Chinese). PubMed/NCBI | |
|
Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, et al: An IAP-IAP complex inhibits apoptosis. J Biol Chem. 279:34087–34090. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Singh R, Letai A and Sarosiek K: Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 20:175–193. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Park JS, Deng JH and Bai Y: Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr. 38:283–291. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Schwarzerová K, Bellinvia E, Martinek J, Sikorová L, Dostál V, Libusová L, Bokvaj P, Fischer L, Schmit AC and Nick P: Tubulin is actively exported from the nucleus through the Exportin1/CRM1 pathway. Sci Rep. 9:57252019. View Article : Google Scholar : PubMed/NCBI | |
|
Yang YL and Li XM: The IAP family: Endogenous caspase inhibitors with multiple biological activities. Cell Res. 10:169–177. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Jo MJ, Jeong S, Yun HK, Kim DY, Kim BR, Kim JL, Na YJ, Park SH, Jeong YA, Kim BG, et al: Genipin induces mitochondrial dysfunction and apoptosis via downregulation of Stat3/mcl-1 pathway in gastric cancer. BMC Cancer. 19:7392019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen M, Tong C, Wu Q, Zhong Z, He Q, Zeng L and Xiao L: 6-Shogaol inhibits the cell migration of colon cancer by suppressing the EMT process through the IKKβ/NF-κB/Snail pathway. Integr Cancer Ther. 22:153473542311727322023. View Article : Google Scholar : PubMed/NCBI | |
|
Li X, Wei Y and Wei X: Napabucasin, a novel inhibitor of STAT3, inhibits growth and synergises with doxorubicin in diffuse large B-cell lymphoma. Cancer Lett. 491:146–161. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Dong H, Hu L, Li W, Shi M, He L, Wang C, Hu Y, Wang H, Wen C, Liu H and Yang X: Pyrimethamine inhibits cell growth by inducing cell senescence and boosting CD8+ T-cell mediated cytotoxicity in colorectal cancer. Mol Biol Rep. 49:4281–4292. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Murugesan A, Lassalle-Claux G, Hogan L, Vaillancourt E, Selka A, Luiker K, Kim MJ, Touaibia M and Reiman T: Antimyeloma potential of caffeic acid phenethyl ester and its analogues through Sp1 mediated downregulation of IKZF1-IRF4-MYC axis. J Nat Prod. 83:3526–3535. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Sari C, SÜmer C and Celep EyÜpoĞlu F: Celep EyÜpoĞlu, Caffeic acid phenethyl ester induces apoptosis in colorectal cancer cells via inhibition of survivin. Turk J Biol. 4:264–274. 2020. View Article : Google Scholar | |
|
Kleczka A, Kubina R, Dzik R, Jasik K, Stojko J, Cholewa K and Kabała-Dzik A: Caffeic acid phenethyl ester (CAPE) induced apoptosis in serous ovarian cancer OV7 cells by deregulation of BCL2/BAX genes. Molecules. 25:35142020. View Article : Google Scholar : PubMed/NCBI | |
|
Colombo D, Gatti L, Sjöstrand L, Carenini N, Costantino M, Corna E, Arrighetti N, Zuccolo M, De Cesare M, Linder S, et al: Caffeic acid phenethyl ester targets ubiquitin-specific protease 8 and synergizes with cisplatin in endometrioid ovarian carcinoma cells. Biochem Pharmacol. 197:1149002022. View Article : Google Scholar : PubMed/NCBI | |
|
Marin EH, Paek H, Li M, Ban Y, Karaga MK, Shashidharamurthy R and Wang X: Caffeic acid phenethyl ester exerts apoptotic and oxidative stress on human multiple myeloma cells. Invest New Drugs. 37:837–848. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Pfeffer CM and Singh ATK: Apoptosis: A target for anticancer therapy. Int J Mol Sci. 19:4482018. View Article : Google Scholar : PubMed/NCBI | |
|
Koff JL, Ramachandiran S and Bernal-Mizrachi L: A time to kill: Targeting apoptosis in cancer. Int J Mol Sci. 16:2942–2955. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Fulda S and Debatin KM: Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 25:4798–811. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Choi SJ, Ahn CH, Hong KO, Kim JH, Hong SD, Shin JA and Cho SD: Molecular mechanism underlying the apoptotic modulation by ethanol extract of Pseudolarix kaempferi in mucoepidermoid carcinoma of the salivary glands. Cancer Cell Int. 21:4272021. View Article : Google Scholar : PubMed/NCBI | |
|
Chung TW, Choi H, Lee JM, Ha SH, Kwak CH, Abekura F, Park JY, Chang YC, Ha KT, Cho SH, et al: Oldenlandia diffusa suppresses metastatic potential through inhibiting matrix metalloproteinase-9 and intercellular adhesion molecule-1 expression via p38 and ERK1/2 MAPK pathways and induces apoptosis in human breast cancer MCF-7 cells. J Ethnopharmacol. 195:309–317. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Cheng AC, Jian CB, Huang YT, Lai CS, Hsu PC and Pan MH: Induction of apoptosis by Uncaria tomentosa through reactive oxygen species production, cytochrome c release, and caspases activation in human leukemia cells. Food Chem Toxicol. 45:2206–2218. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Roy S and Nicholson DW: Cross-talk in cell death signaling. J Exp Med. 192:F21–F25. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Gavrilescu LC and Denkers EY: Apoptosis and the balance of homeostatic and pathologic responses to protozoan infection. Infect Immun. 71:6109–6115. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Suzuki S, Yamamoto M, Sanomachi T, Togashi K, Sugai A, Seino S, Yoshioka T, Kitanaka C and Okada M: Brexpiprazole, a serotonin-dopamine activity modulator, can sensitize glioma stem cells to osimertinib, a third-generation EGFR-TKI, via survivin reduction. Cancers. 11:9472019. View Article : Google Scholar : PubMed/NCBI | |
|
Sakoguchi-Okada N, Takahashi-Yanaga F, Fukada K, Shiraishi F, Taba Y, Miwa Y, Morimoto S, Iida M and Sasaguri T: Celecoxib inhibits the expression of survivin via the suppression of promoter activity in human colon cancer cells. Biochem Pharmacol. 73:1318–1329. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Dean E, Jodrell D, Connolly K, Danson S, Jolivet J, Durkin J, Morris S, Jowle D, Ward T, Cummings J, et al: Phase I trial of AEG35156 administered as a 7-day and 3-day continuous intravenous infusion in patients with advanced refractory cancer. J Clin Oncol. 27:1660–1666. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Yue C, Li RH, Chen C and Liu H: Study on the relationship between XIAP gene and resistance of taxol in ovarian cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 49:337–341. 2018.(In Chinese). PubMed/NCBI | |
|
Hehlgans S, Petraki C, Reichert S, Cordes N, Rödel C and Rödel F: Double targeting of Survivin and XIAP radiosensitizes 3D grown human colorectal tumor cells and decreases migration. Radiother Oncol. 108:32–39. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Werner TA, Dizdar L, Nolten I, Riemer JC, Mersch S, Schütte SC, Driemel C, Verde PE, Raba K, Topp SA, et al: Survivin and XIAP-two potential biological targets in follicular thyroid carcinoma. Sci Rep. 7:113832017. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Gao W, Ma Y, Zhu G, Chen F and Qu H: Dual targeting of survivin and X-linked inhibitor of apoptosis protein suppresses the growth and promotes the apoptosis of gastric cancer HGC-27 cells. Oncol Lett. 16:3489–3498. 2018.PubMed/NCBI | |
|
Fang W, Che X, Li G, Wang A, Wang Y, Shi X, Hou K, Zhang X, Qu X and Liu Y: Sur-X, a novel peptide, kills colorectal cancer cells by targeting survivin-XIAP complex. J Exp Clin Cancer Res. 39:822020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu TT, Yang H, Zhuo FF, Yang Z, Zhao MM, Guo Q, Liu Y, Liu D, Zeng KW and Tu PF: Atypical E3 ligase ZFP91 promotes small-molecule-induced E2F2 transcription factor degradation for cancer therapy. EBioMedicine. 86:1043532022. View Article : Google Scholar : PubMed/NCBI | |
|
Dohi T, Xia F and Altieri DC: Altieri, Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol Cell. 27:17–28. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Arora V, Cheung HH, Plenchette S, Micali OC, Liston P and Korneluk RG: Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J Biol Chem. 282:26202–26209. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Hoffman WH, Biade S, Zilfou JT, Chen J and Murphy M: Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 277:3247–3257. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, et al: Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 21:2613–2622. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Baek HS, Kwon YJ, Ye DJ, Cho E, Kwon TU and Chun YJ: CYP1B1 prevents proteasome-mediated XIAP degradation by inducing PKCε activation and phosphorylation of XIAP. Biochim Biophys Acta Mol Cell Res. 1866:1185532019. View Article : Google Scholar : PubMed/NCBI | |
|
Hong SW, Shin JS, Moon JH, Jung SA, Koh DI, Ryu Y, Park YS, Kim DY, Park SS, Hong JK, et al: Chemosensitivity to HM90822, a novel synthetic IAP antagonist, is determined by p-AKT-inducible XIAP phosphorylation in human pancreatic cancer cells. Invest New Drugs. 38:1696–1706. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Chung TW, Moon SK, Chang YC, Ko JH, Lee YC, Cho G, Kim SH, Kim JG and Kim CH: Novel and therapeutic effect of caffeic acid and caffeic acid phenyl ester on hepatocarcinoma cells: Complete regression of hepatoma growth and metastasis by dual mechanism. FASEB J. 18:1670–1681. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Tang H, Yao X, Yao C, Zhao X, Zuo H and Li Z: Anti-colon cancer effect of caffeic acid p-nitro-phenethyl ester in vitro and in vivo and detection of its metabolites. Sci Rep. 7:75992017. View Article : Google Scholar : PubMed/NCBI | |
|
Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V, Ibbs M, Bliźniak R, Łuczewski Ł and Lamperska K: 2D and 3D cell cultures-a comparison of different types of cancer cell cultures. Arch Med Sci. 14:910–919. 2018.PubMed/NCBI | |
|
Mañós M, Giralt J, Rueda A, Cabrera J, Martinez-Trufero J, Marruecos J, Lopez-Pousa A, Rodrigo JP, Castelo B, Martínez-Galán J, et al: Multidisciplinary management of head and neck cancer: First expert consensus using Delphi methodology from the Spanish Society for Head and Neck Cancer (part 1). Oral Oncol. 70:58–64. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Kuo YY, Lin HP, Huo C, Su LC, Yang J, Hsiao PH, Chiang HC, Chung CJ, Wang HD, Chang JY, et al: Caffeic acid phenethyl ester suppresses proliferation and survival of TW2.6 human oral cancer cells via inhibition of Akt signaling. Int J Mol Sci. 14:8801–8817. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Peng CY, Yang HW, Chu YH, Chang YC, Hsieh MJ, Chou MY, Yeh KT, Lin YM, Yang SF and Lin CW: Caffeic Acid phenethyl ester inhibits oral cancer cell metastasis by regulating matrix metalloproteinase-2 and the mitogen-activated protein kinase pathway. Evid Based Complement Alternat Med. 2012:7325782012. View Article : Google Scholar : PubMed/NCBI | |
|
Machiels JP, René Leemans C, Golusinski W, Grau C, Licitra L and Gregoire V; EHNS Executive Board, ESMO Guidelines Committee, : ESTRO Executive Board: Reprint of ‘Squamous cell carcinoma of the oral cavity, larynx, oropharynx and hypopharynx: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up’. Ann Oncol. 31:1462–1475. 2020. View Article : Google Scholar : PubMed/NCBI |